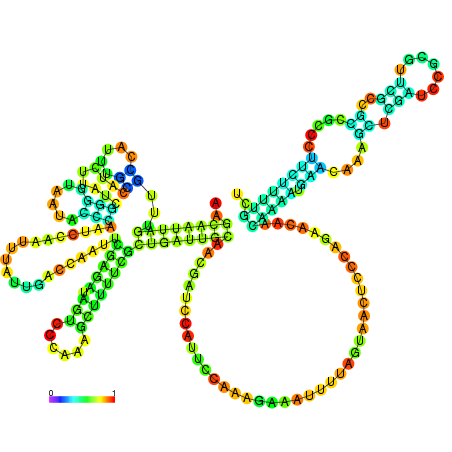| dm3 |
chr2R:18872718-18872857 - |
ATGCACATTTATTTGGCATCATG------------------------------------------C---------------------------------------TTCTGCGATCTCTACCAATTTTCCTTGCACTCT------TTGTCCTTTCCAAAAGTGACACGGGT----GCAGGTGAAGATTCG---GAAGAGGACGACGAAAACGACT---GCAAC-CGGACGACTCTTGGT |
| droSim1 |
chr2R:17459524-17459663 - |
ATCCACATTTATTTGGCATCATG------------------------------------------C---------------------------------------TTCTGCGAGCTCTACCACTTCTCCTTGCACTCT------TTGTCCTTTCCAAAAGCGACACTGAT----GCAGGTGAAGATTCG---GAGGAGGACGACAGCGAAGACT---GCAAC-AGGACGACTCTTGGT |
| droSec1 |
super_9:2164747-2164886 - |
ATCCACATTTATTTGGCATCATG------------------------------------------C---------------------------------------TTCTGCGAGCTCTACCACTTCTCCTTGCACTCT------TTGTCCTTTCCAAAAGCGACACTGAT----GCAGGTGAAGATTCA---GAGGAGGACGACAGCGAAGACT---GCAAC-AGGACGACTCTTGGT |
| droYak2 |
chr2R:18832811-18832950 - |
AACCGCGTTTATTTGCCAAAATG------------------------------------------C---------------------------------------TGCTCCGAGCTCTACCACTTCTGCTCACACTCTGTGTGTTGGTCCTCTCCAACGACGACACGGAT----GCAGATGAAGAAGAG---GAAGAGGACGA------CGACT---GCAAC-AAGGCGACTCTTGGT |
| droEre2 |
scaffold_4845:20234099-20234238 - |
AACCACGTTTATTTGCCATAATG------------------------------------------C---------------------------------------TGCTCCGAACTCTGCTC------CTTACGCTCTTCGTGTTTGTCCTTGCCAAAAACGATACGATT----GCAGAAGTAGAGGAA---GAGTCGGACGAGGACGAAGGAT---GCAAC-AAGGCGACTCTTGGT |
| droAna3 |
scaffold_13266:15204126-15204261 - |
ATTTAAGA-GCTTTTCCCTTCAAAGCCT----------------------------------------------------------------------------------------------AAACAGTTGACACAAAATGTGGCTACCCTTCTTGCTAGTGGCACTGCC----TTGGGGGACTGTGGTGGG---GGGAGGTCCTCTGACATTGGCCAATCTTAGCCGAATCATTGGA |
| dp4 |
chr3:14089101-14089284 - |
AAGCA-ATTAGTTTGCCATTTGCCATCTTATCGGGGTAATACCCAATCCAATTTATTGACCAATTCGAGATAGTCCCAAAGCTTTTCGCTGATTGCAACGATCCATTCCAAAGAAATTTT----------------------------------------------AGTAACTCCCAGAACAACAAAATGAA---CAAAGCTCGATCCGCGTTC---GCCGC-CGCCCTTCTTTTGCT |
| droPer1 |
super_4:1228949-1229132 + |
AAGCA-ATTAGTTTGCCATTTGCCATCTTATCGGGGTAATACCCAATCCAATTTATTGACCAATTCAAGATAGTCCCAAAGCTTTTCGCTGATTGCAACGATCCATTCCAAAGAAATTTT----------------------------------------------AGTAGCTCCCAGAACAACAAAATGAA---CAAAGCTCGATCCGCGTTC---GCCGC-CGCCCTTCTTTTGCT |
| droWil1 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6496:24343095-24343103 - |
AT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTTGGC |
| droGri2 |
scaffold_15112:3934700-3934745 - |
ATATTCAATATGCT-------------------------------------------GAGCACATC---------------------------------------TTCTGAAAGCATTTCAGACTTTC-------------------------------------------------------------------------------------------------------------- |