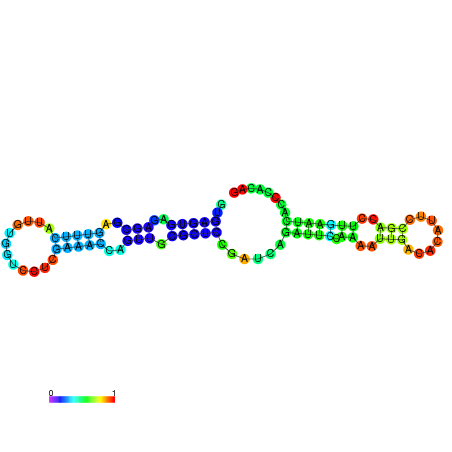
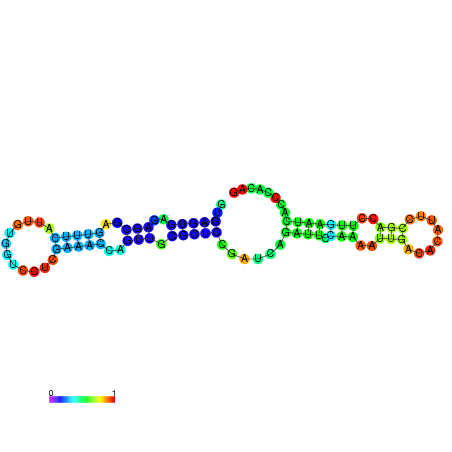
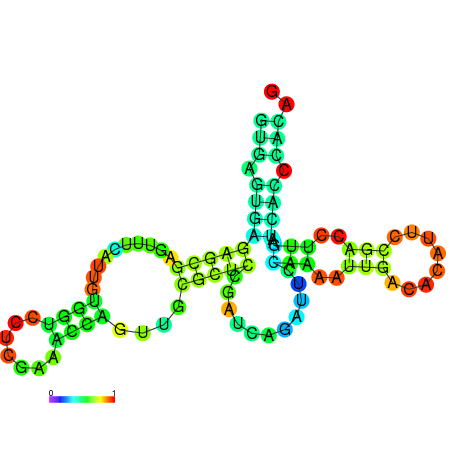
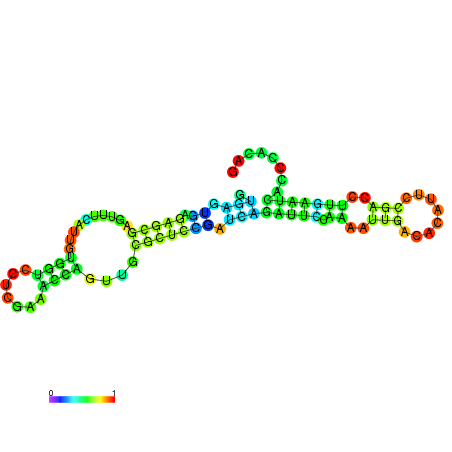
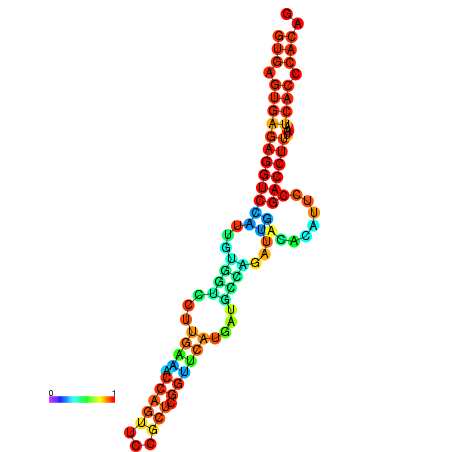
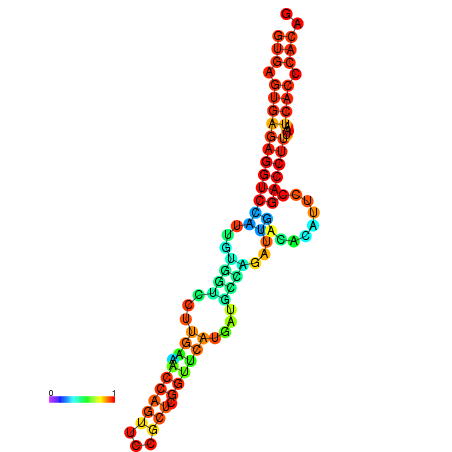
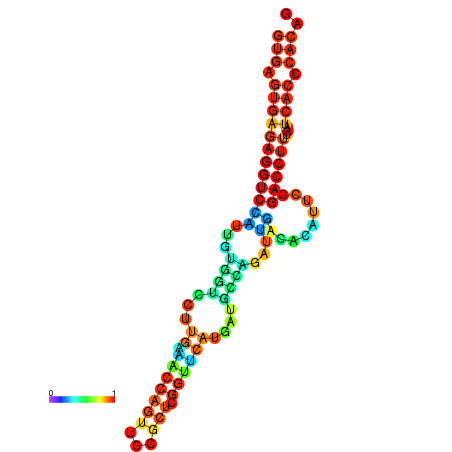
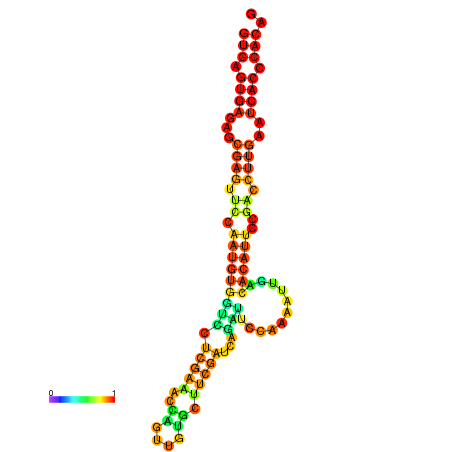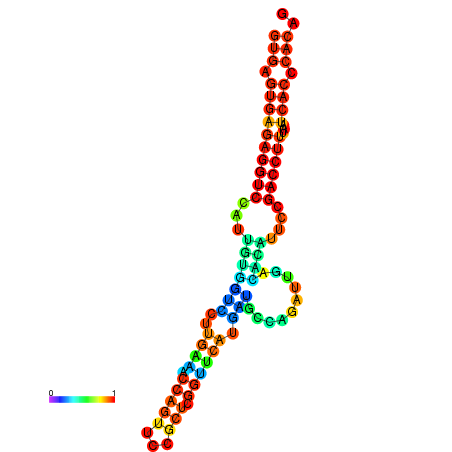| dm3 |
chr2L:2737553-2737676 + |
AGACGAGCCCACAACGTGAGTGA-GAGCGAGT------TCCAAT-----------------------GTGGTCC----------------------TCGAAACCAGTTGTGCTTCGATCAGATTCCAAAATTGACACATT--CCG---------------------------------ACCTTGAATCA-CCCACAGCGGATTGGACAGCTC |
| droSim1 |
chr2L:2697912-2698035 + |
AGACGAGCCCACAACGTGAGTGA-GAGCGAGT------TTCATT-----------------------GTGGTCC----------------------TCGAAACCAGTTGCGCTCCGATCAGATTCCAAAATTGACACATT--CCG---------------------------------ACCTTGAATCA-CCCACAGCGGCTTGGACAGCTC |
| droSec1 |
super_5:903635-903758 + |
AGACGAGCCCACAACGTGAGTGA-GAGCGAGT------TCCATT-----------------------GTGGTCC----------------------TCGAAACCAGTTGCGCTCCGATCAGATTCCAAAATTGACACATT--CCG---------------------------------ACCTTGAATCA-CCCACAGCGGCTTGGACAGCTC |
| droYak2 |
chr2L:2732711-2732833 + |
AGACGAGCCCACAACGTGAGTGA-GA----------GGTCCATT-----G-----------T-----GTGATCC----------------------CCAAAACCAGTTCTACTCAGATCTGATTCC-AAATTGACACATTCTCTG---------------------------------ACCTTGAATCA-CCCACAGCGGATTGGATAGCTC |
| droEre2 |
scaffold_4929:2790150-2790269 + |
AGACGAGCCCACAACGTGAGTGA-GA----------GGTCCATT-----------------------GTGGTCC----------------------TTGAAACCAGTTCCGCTCGGTTCATGATGCCAGATTGACACATT--CCG---------------------------------ACCTTTAATCA-CCCACAGCGGATTGGACAGCTC |
| droAna3 |
scaffold_12916:13439490-13439581 + |
GGACGAGCCAACAACGTAAGT--------------------------------------------------TCT----------------------CTGAAACCATTC----------CAATTTTCAAACTTATCAAGTT--G-----------------------------------TTAATGTCTTT-CACACAGGGGCTTGGATAGTTC |
| dp4 |
chr4_group4:2073904-2074060 + |
AGACGAGCCCACAACGTAAGTATCGAACGAGTGAGAGGACCA-GACCAG--AGGCAGCATCTGCGATGCGGTCGAAAAGAGAAGAGAAACCAGAGAGCTAAACCAGTTGCTGACT-----------------GACATTCC--ACA---------------------------------ACCTTGAACCATCCCACAGAGGCCTGGATAGTTC |
| droPer1 |
super_10:1078988-1079144 + |
AGACGAGCCCACAACGTAAGTATCGAACGAGTGAGAGGACCA-GACCAG--AGGCAGCATCTGCGATGCGGTCGAAAAGAGAAGAGAAACCAGAGAGCTAAACCAGTTGCTGACT-----------------GACATTCC--ACA---------------------------------ACCTTGAACCATCCCACAGAGGCCTGGATAGTTC |
| droWil1 |
scaffold_180772:1508923-1509021 + |
AGACGAGCCCACAACGTAAGTAA-TAACAAA------------------------------------------------------------------AAAGACCGGTTATTTCCT-----------------G---------TTAGCCTTGAACTTAACCAACATCTC-------------------AA-CCAACAGAGGCCTGGACAGCTC |
| droVir3 |
scaffold_12963:11356884-11356997 + |
AGACGAGCCCACAACGTAAGCAA-GAATAGAG------A------ATGCAATCAAT-----------G----------------------------CGGAAACCAGTTGCTGACT-----------------AACATGTG--CTTACCTTGAAATTG-----T---------------------AACCA-TTCGCAGGGGCCTGGATAGTTC |
| droMoj3 |
scaffold_6308:1548466-1548588 - |
CGACGAGCCCACGTCGGGCCTGG-------A-----C-TCCTTTACCGC--CCACAGC---------------------------------------GTC---------------GTCCAGGTGCTGAAGAAGCTA----------TCGCAAAAGGG-----CAAGACCGTCATACTGACCATACATCA-GCCATCG----TCGGAGCTCTT |
| droGri2 |
scaffold_15252:5474360-5474483 + |
GGACGAGCCCACAACGTAAGTGA-AA----------CA-CCGAAACACCA-----------C-----ACAATC------------------------GAGAACCAGTTACTGACT----AA------GCATTATTATTTT--TTAACCTTGAAATTG-----CATCGC------------------ATC-CATACAGGGGCCTGGACAGTTC |