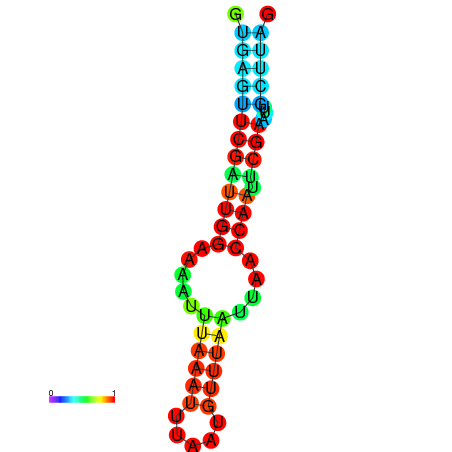
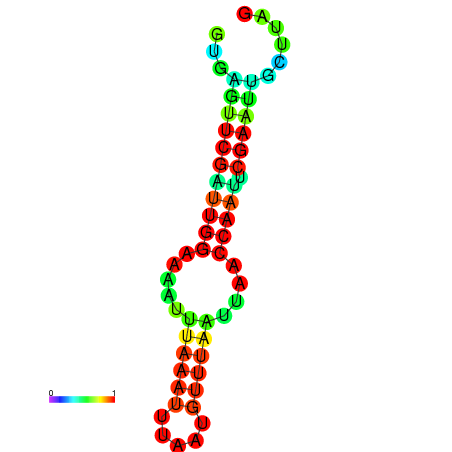
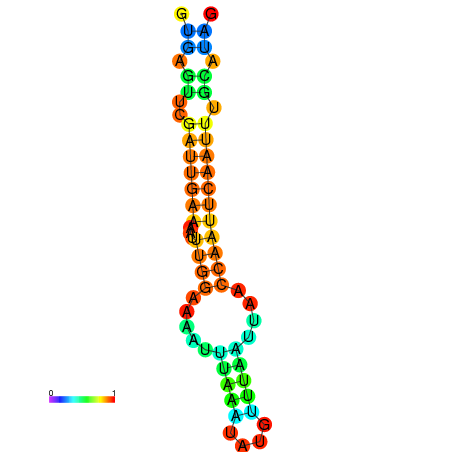
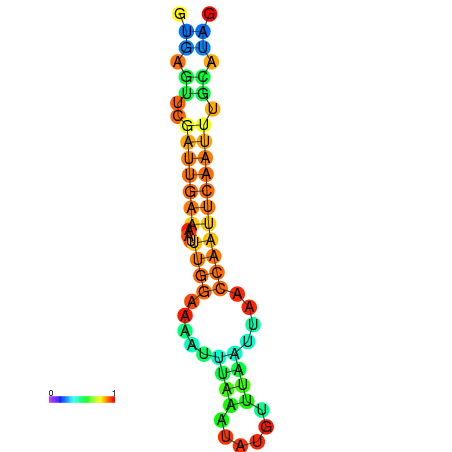
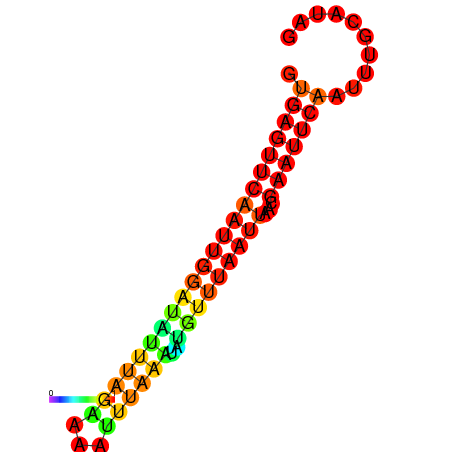
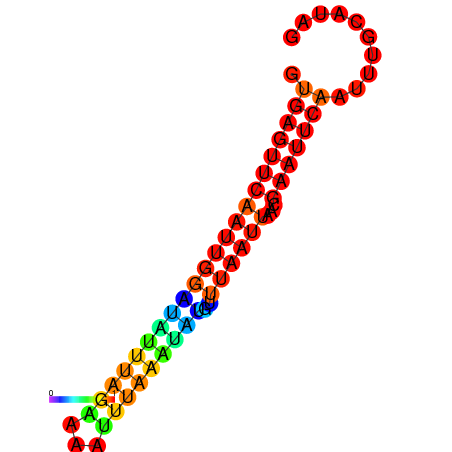
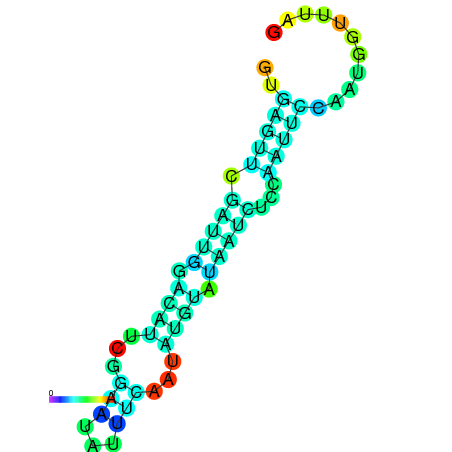
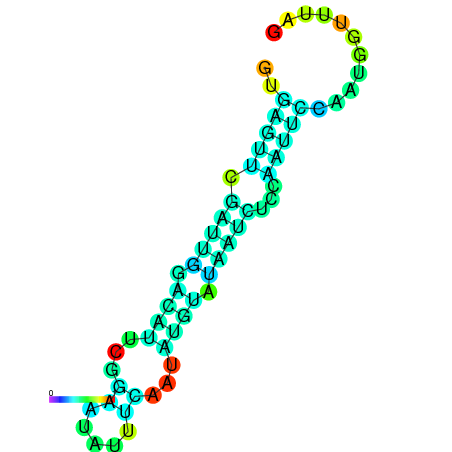
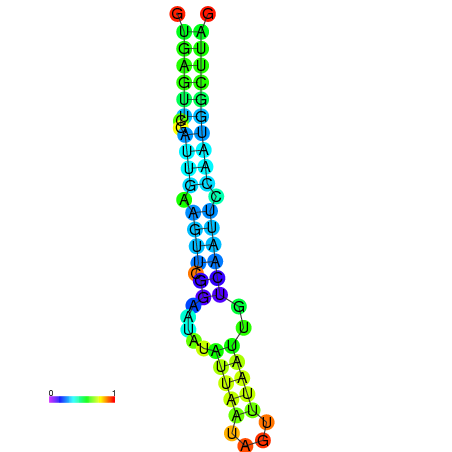
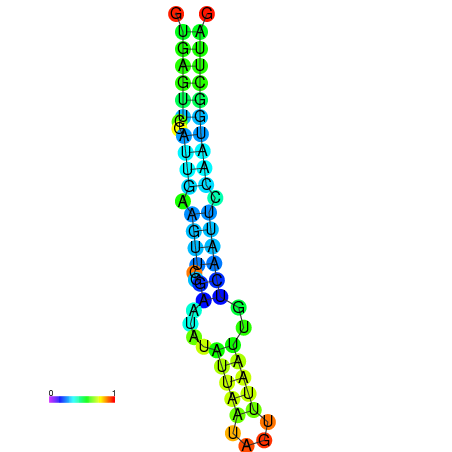
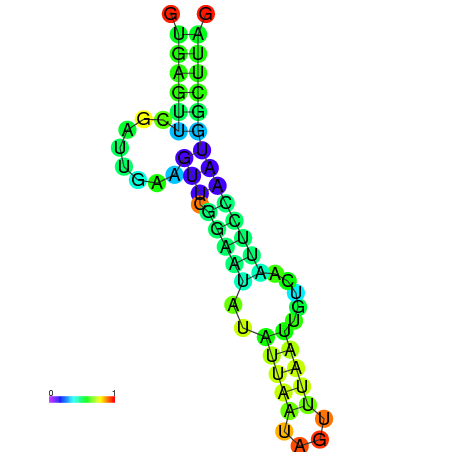
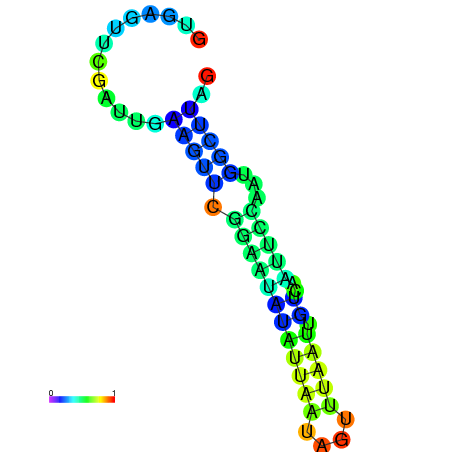
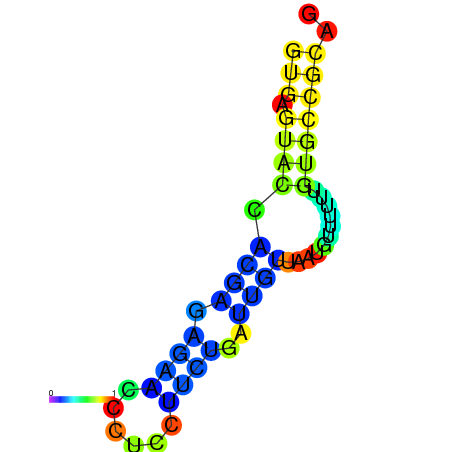
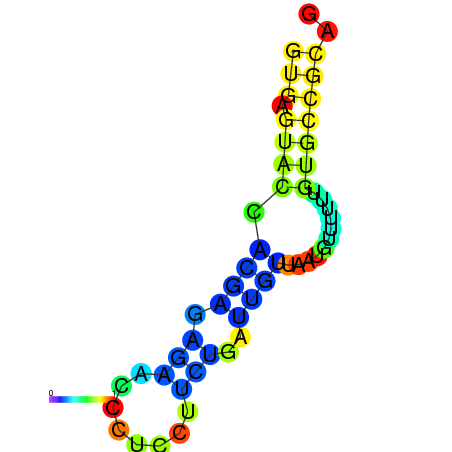
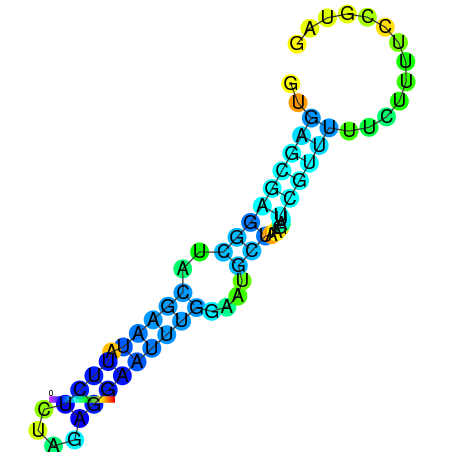
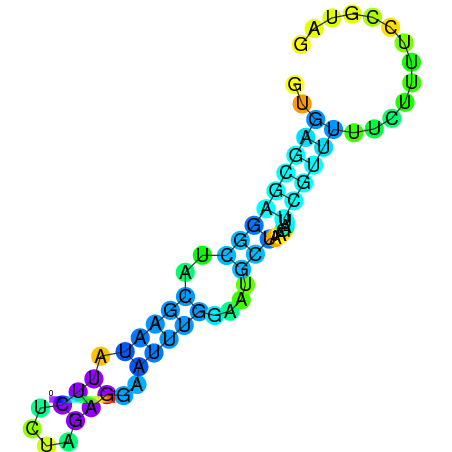
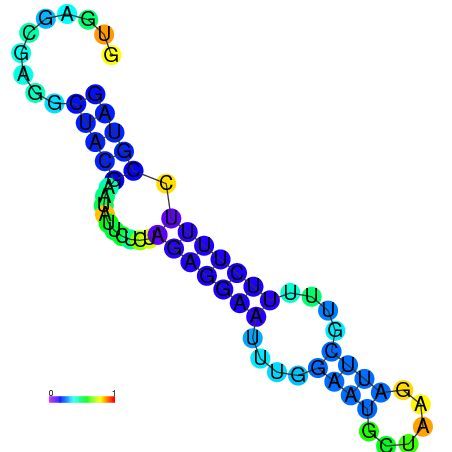
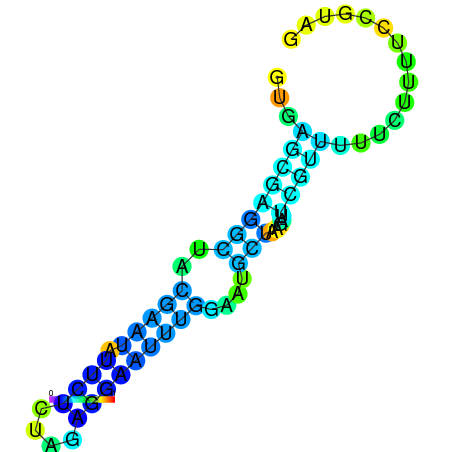
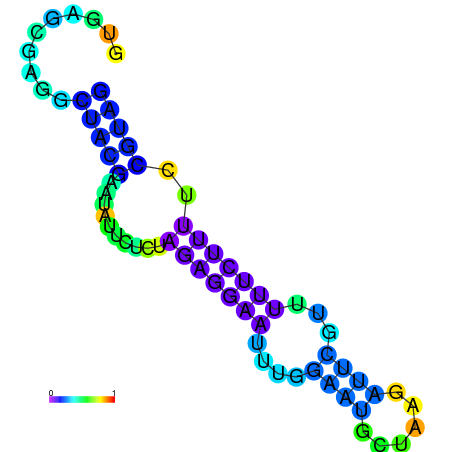
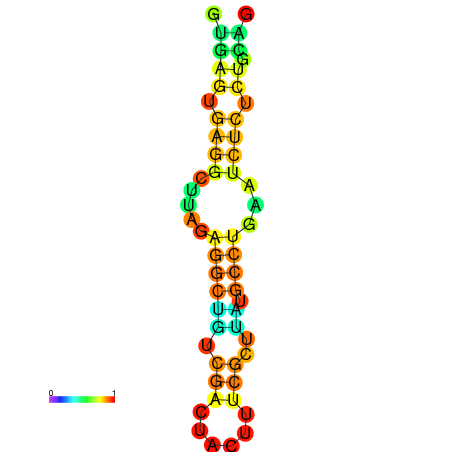
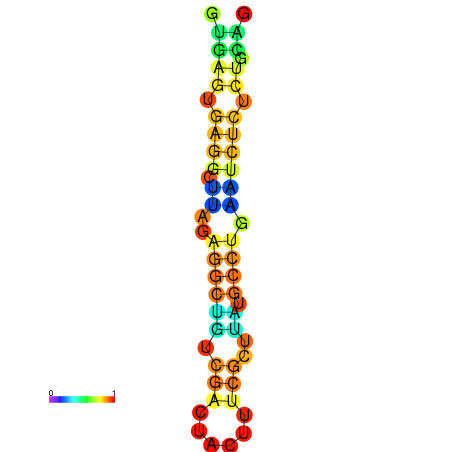
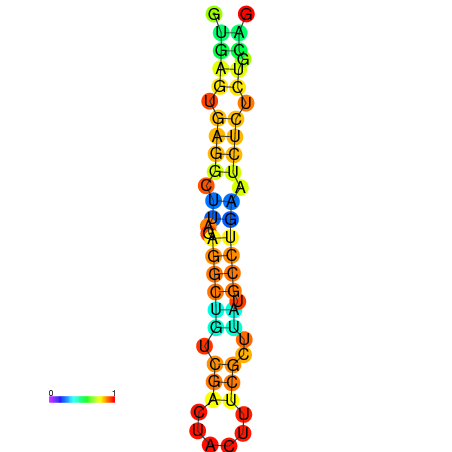
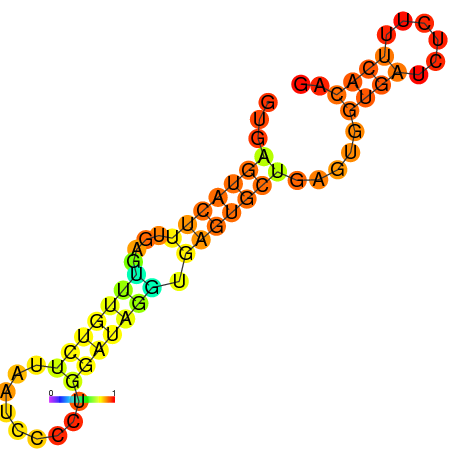
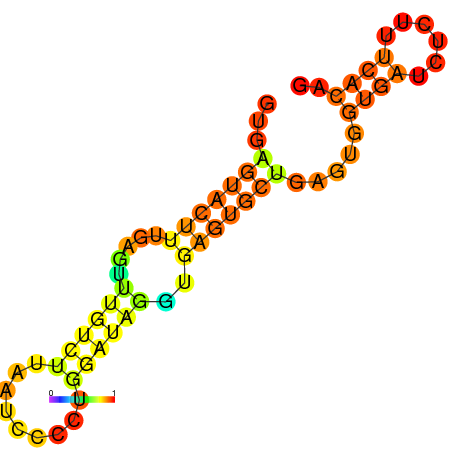
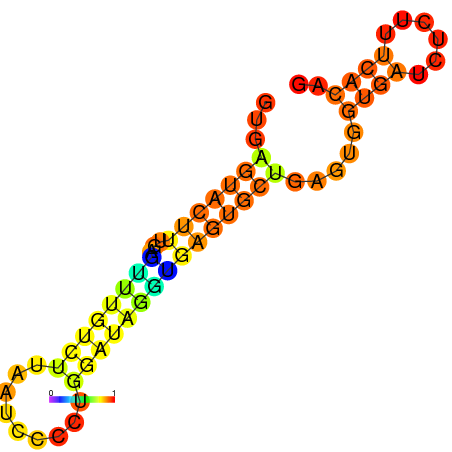
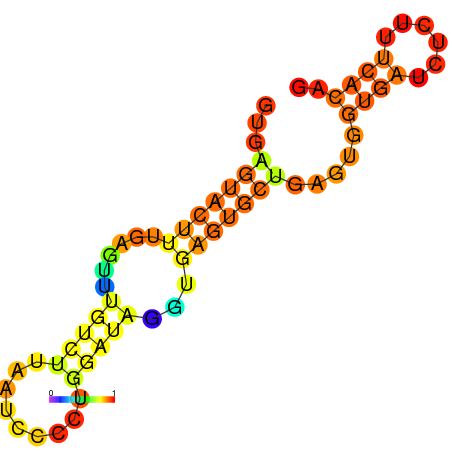
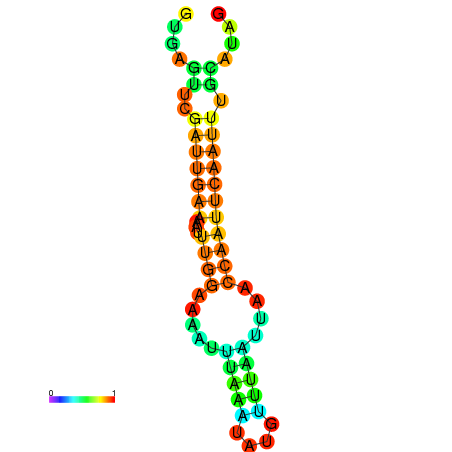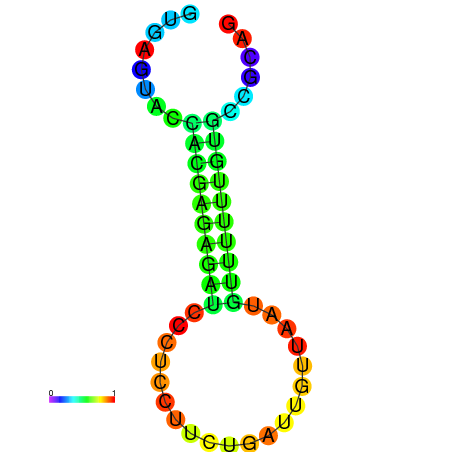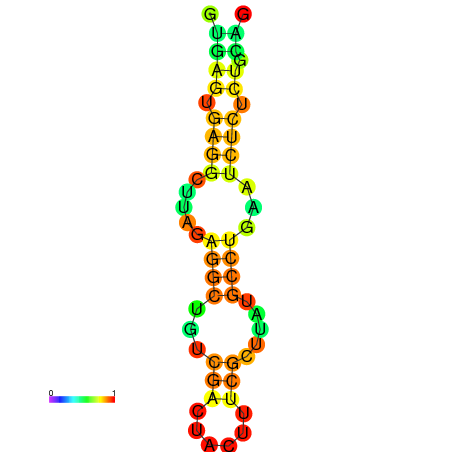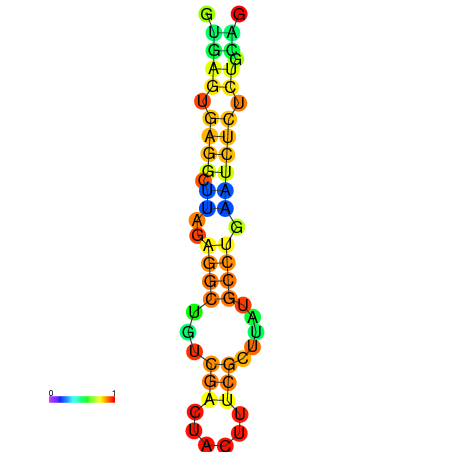| dm3 |
chr3L:8347269-8347356 - |
TGTTCAACTACTTGAGTGAGTTCGATT---GGAA-------------------------------AAT---------TTAAATTTAA----TGT---TTAATTAACCAATTCGAATTGCTTAGATTGCTATATTGGCT |
| droSim1 |
chr3L:7807265-7807357 - |
TATTCAACTATTTGAGTGAGTTCGATT---GAAA---------------------AT--TTGGAAAAT---------TTAA-------ATATGT---TTAATTAACCAATTCAATTTGCATAGATTGCTACATTGGCT |
| droSec1 |
super_0:643746-643838 - |
TATTCAACTATTTGAGTGAGTTCAATT---GGAT---------------------AT--TTAGAAAAT---------TTAA-------ATATGT---TTAATTAACGAATTCAATTTGCATAGATTGCTACATTGGCT |
| droYak2 |
chr3L:3551197-3551288 + |
TATTCAACTACTTGAGTGAGTTCGATT---GGAC---------------------AT--TCGGAATAT---------TTCA-------ATATGT---ATAATCT-CCAATTCCAATGGTTTAGATTGCTACATTGGCT |
| droEre2 |
scaffold_4784:5683607-5683696 + |
TATTCAACTATTTGAGTGAGTTCGATT---GAA----------------------GT--TCGGAATAT---------ATTA-------ATA-GT---TTAATTGT-CAATTCCAATGGCTTAGATTGCTACATTGGCT |
| droAna3 |
scaffold_13337:7445991-7446073 + |
TGTTCAACTATTTGAGTGAGTATCTTC---CTA------------------AGAAGGAAGT-------GTC--AT--T---TATTTA----TGAT---------------A-TTACTGCATAGATTGCTACATCGGCT |
| dp4 |
chrXR_group6:12821045-12821128 - |
TTTTCAACTATTTGAGTGAGTACCACG---AGAGAT-----------------------CC-------CTC--CTTCTGATTGTTAA----TGTT---------------TTTTGTGCCGCAGATTGCTATATCGGCT |
| droPer1 |
super_33:845496-845579 - |
TTTTCAACTATTTGAGTGAGTACCACG---AGAGAA-----------------------CC-------CTC--CTTCTGATTGTTAA----TGTT---------------TTTTGTGCCGCAGATTGCTATATCGGCT |
| droWil1 |
scaffold_181009:3090042-3090138 + |
TCTTCAATTATTTGAGTGAGCGAGGCTACGA-AT--------ATTCTCTAGAGGAAT--TTGGAATG----------------CTAAGATTCGT---TT-----------TTCTTTTCCGTAGATTGCTACATTGGCT |
| droVir3 |
scaffold_13049:4752940-4753026 - |
TCTTCAACTACTTGAGTGAGTGAGGCTT--AGAGG------------------------CTG-------TCGACTACTTTC-GCTTA----TGCCTG-------------AATCTCTCTGCAGATTGCTATATCGGCT |
| droMoj3 |
scaffold_6680:3143577-3143662 - |
TCTTCAATTATTTGAGTGGGTAGAGTG---AAGTAT-----------------------TT-------CTT--ATACTCCAGCCTAA----TGTTGG-------------TCTCTCCTTATAGATTGCTATATTGGCT |
| droGri2 |
scaffold_15110:10166029-10166124 + |
TCTTCAACTATTTGAGTGAGTACTTTG---AGT-TTGTCTTAATCCCCTGGATAGGTGAG--------------TGCTGAGTGGTGA----TC--------------------TCTTTCACAGATTGCTATATCGGTT |