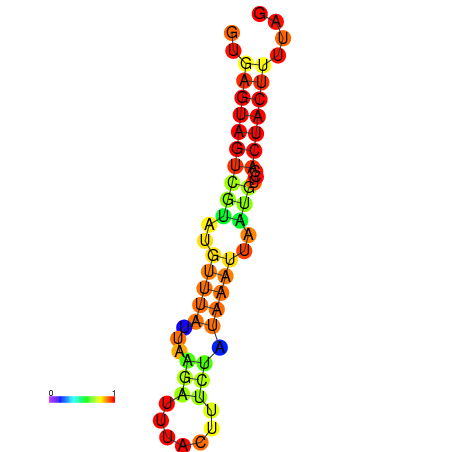
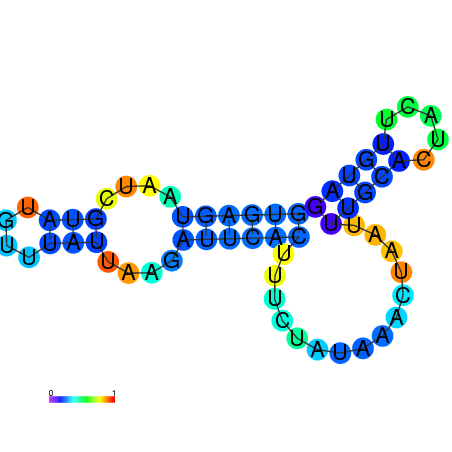
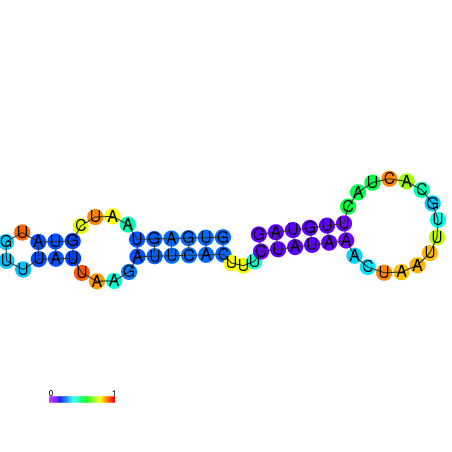
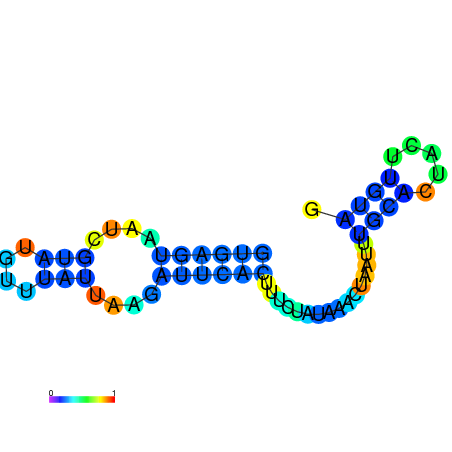
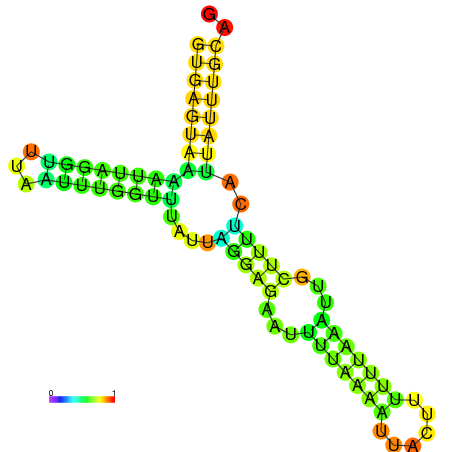
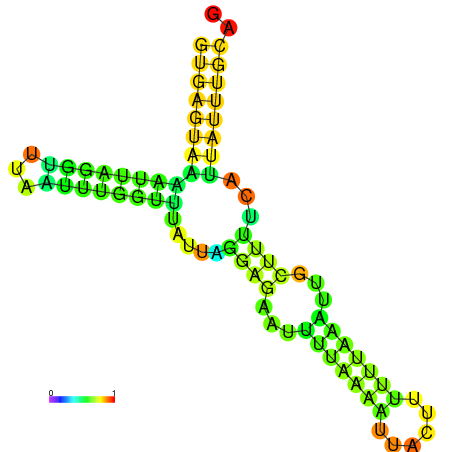
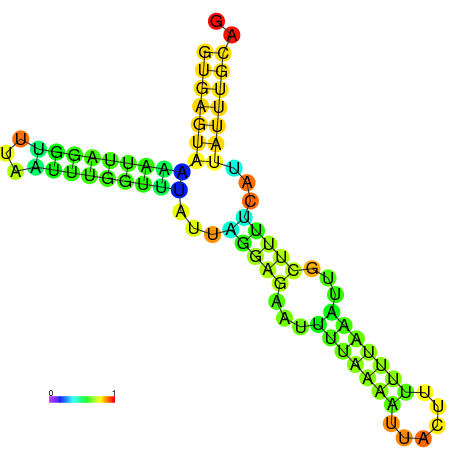
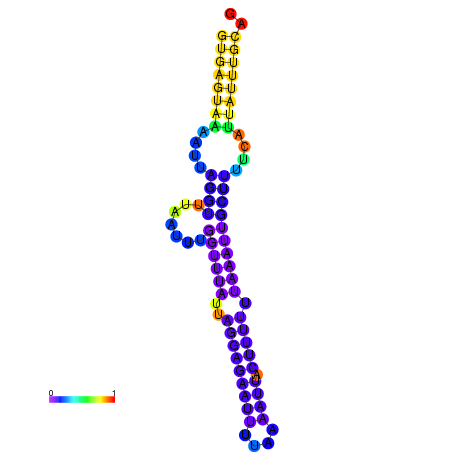
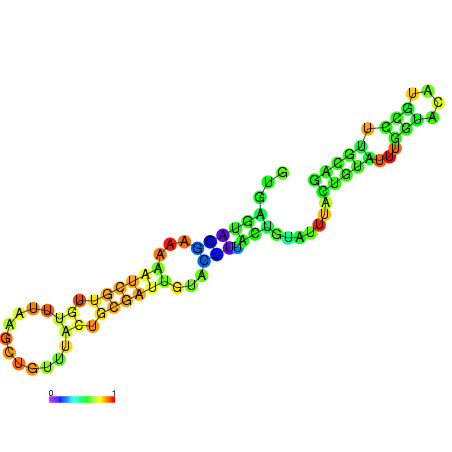
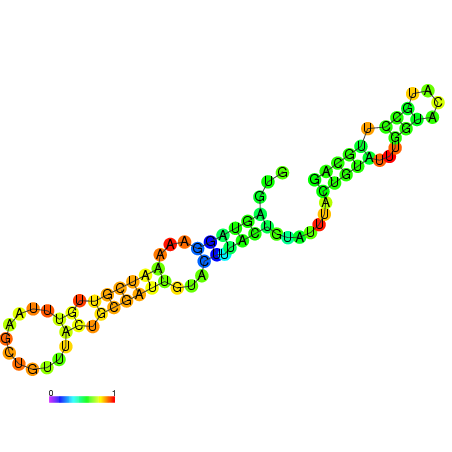
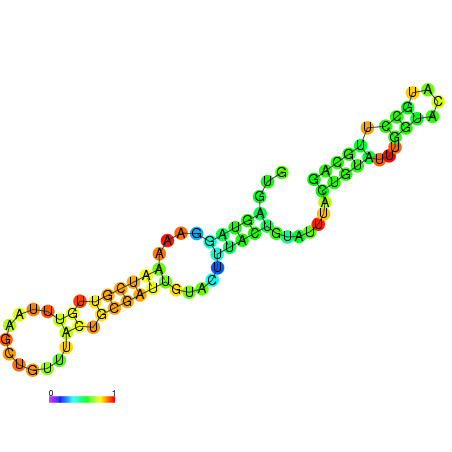
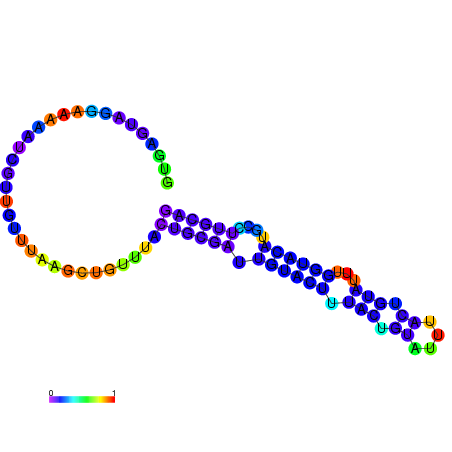
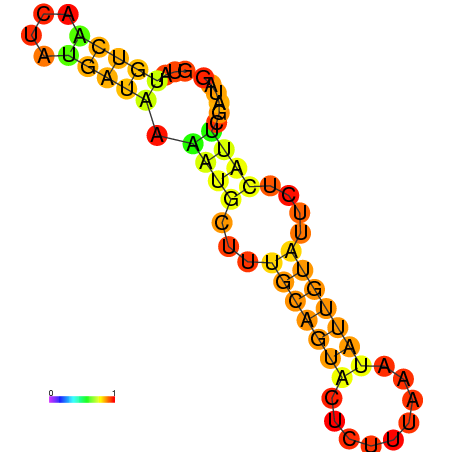
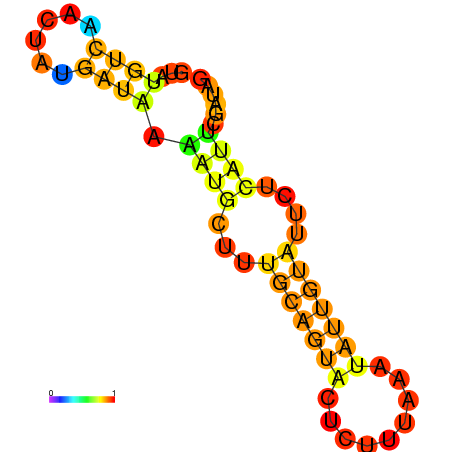
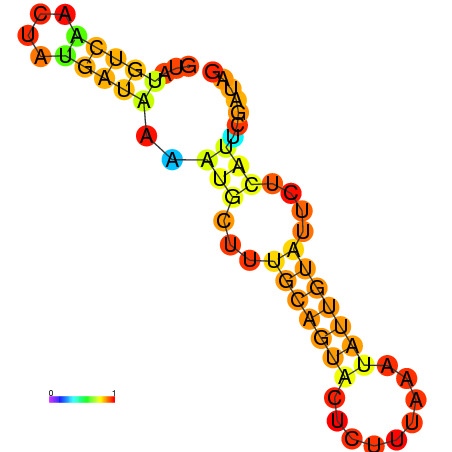
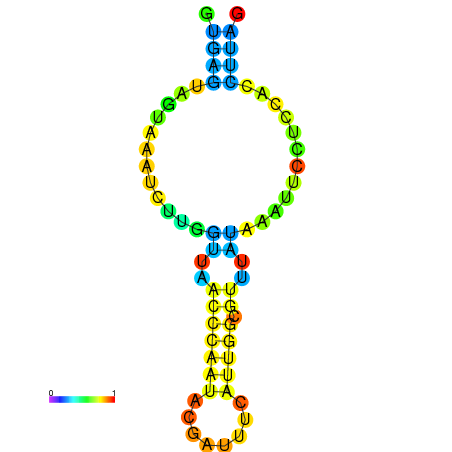| dm3 |
chr2L:4829965-4830054 - |
CATTCGCAGCTTTAAGTGAGTAGTC---G--------------------TATGTTTATTA---------------------AGATT-TACT----TTCTATA--AATTAATGTCCA-----------------------CTACTTTTAGTCTCTATGACGATGA |
| droSim1 |
chr2L:4699797-4699886 - |
CATTCGCAGCTTTAAGTGAGTAATC---G--------------------TATGTTTATTA---------------------AGATT-CACT----TTCTATA--AACTAATTTGCA-----------------------CTACTTGTAGTCTCTATGACGACGA |
| droSec1 |
super_5:2925673-2925762 - |
CATTCGCAGCTTTAAGTGAGTAATC---G--------------------TATGTTTATTA---------------------AGATT-CACT----TTCTATA--AACTAATTTGCA-----------------------CTACTTGTAGTCTCTATGACGACGA |
| droYak2 |
chr2L:4839794-4839887 - |
CATTCGCAGCTTTAAGTGAGTAAAA---C--------------------TATGTCTATTA----------GGTG-------AAATT-AACT----TTCTATA--AATTACTTTTCT-----------------------TTATTTTTAGTCTATATGACGACGA |
| droEre2 |
scaffold_4929:4910720-4910828 - |
CATTCGCAGTTTTAAGTGAGTAAAA---T--------------------TAGGTTTAATTTGGTTTATTAGGAGAATTTTAAAAT--TAC-----TTTTTTA--AATTGCTTTTCA-----------------------TTATTTGCAGTCTCTATGACGACGA |
| droAna3 |
scaffold_12916:9399289-9399374 - |
CATTCGAAGCTTCAAGTATGTTAAGA---------AATCCACGATT-----------------------------------TTT--T-TATATTTTCATACA--AAAT-----TC------------------------ATTTTTTCAGCTTGTACGACGACGA |
| dp4 |
chr4_group3:3587822-3587933 - |
CATACGCACATTTAAGTGAGTAGGAA---------AAATCGTTGTTTAAGCTGTTTACTG---------------------CGATTGTA--------------------CTTTACTGTATTTACTGTATTTGGTA--CATGCCTTGCAGCCTCTATGATTCCGA |
| droPer1 |
super_1:5067398-5067508 - |
CATACGCACATTTAAGTGAGTAGGA----------AAATCGCTGTTTAAGCTGTTTACTG---------------------CGATTGTA--------------------CTTTACTGTATTTACTGTATTTGGTA--CATGCCTTGCAGCCTCTATGATTCCGA |
| droWil1 |
scaffold_180772:1562219-1562313 - |
CATTAAAACGTTTAAGTGAGTAGTAAATCTTGGTTAAC----------------CCAATA---------------------CGATTT-CATTGGCGTTTATA--AATT-----CC------------------------TCCACCTTAGCCTTTACGACGACGA |
| droVir3 |
scaffold_12963:8507047-8507138 + |
CATACGCACATTCAAGTATGTCA---------------------------------ACTA---------------------TGAT----------------A--AAATGCTTTGCAGTACTCTTTAAATATTGTATTCTCATTCGATAGCCTCTATGACGACGA |
| droMoj3 |
scaffold_6500:1118880-1118971 + |
CATACGTACATTCAAGTAAGCCCAA---T-TCGCC-------GACG-----------------------------------TTA--T-GGTATTCATATACAAATGCTAATCATTT-----------------------ATCCCTTTAGCCTCTATGACGACGA |
| droGri2 |
scaffold_15252:11699277-11699378 + |
CATACGCACCTTCAAGTATGTGCAA---T-GGGTAAACCATATGTGAGAGATTTAAACCT---------------------TAATTTTA--------------------CTTTCCTATATCT-----------------GCCCAAACAGTCTGTACGATGACGA |