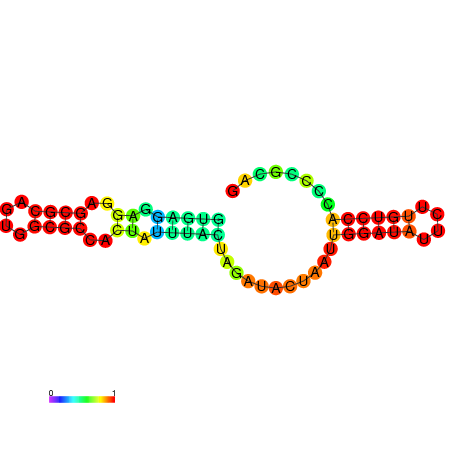
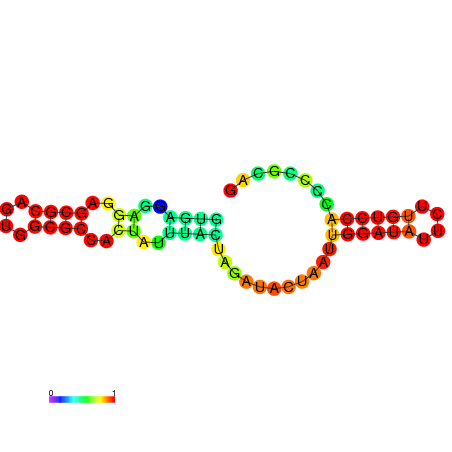
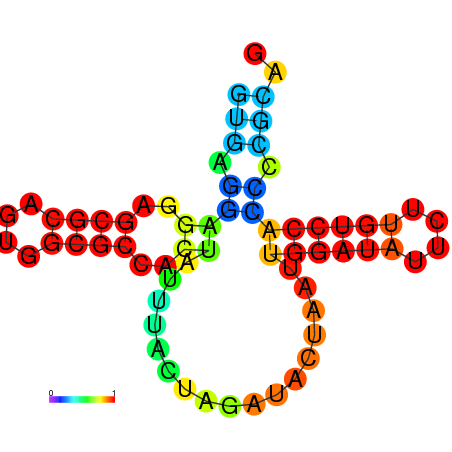
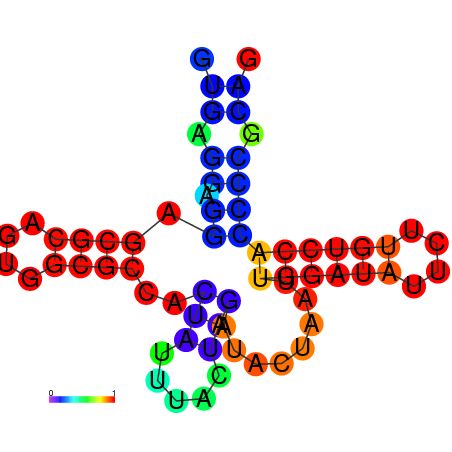
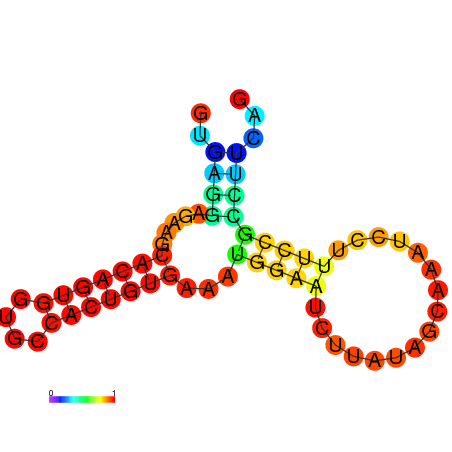
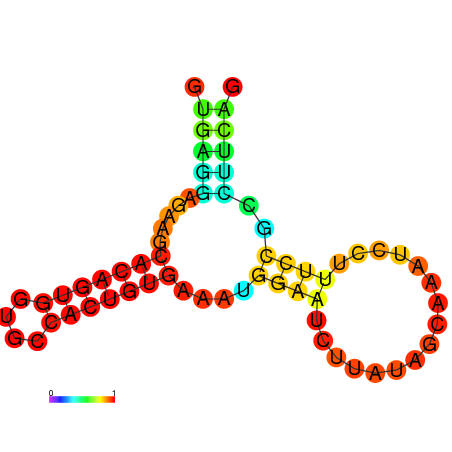
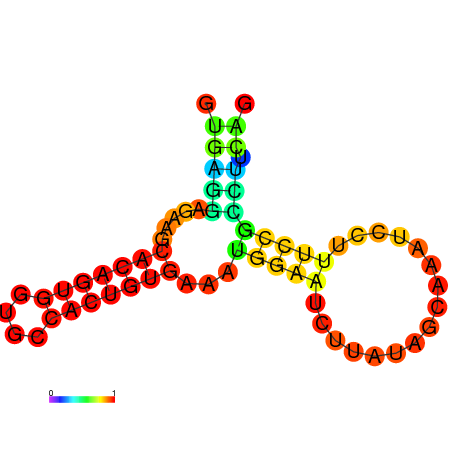
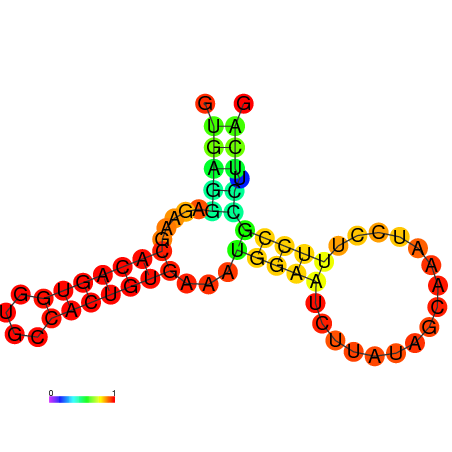
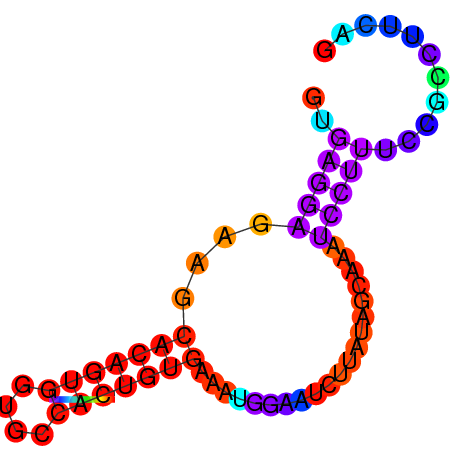
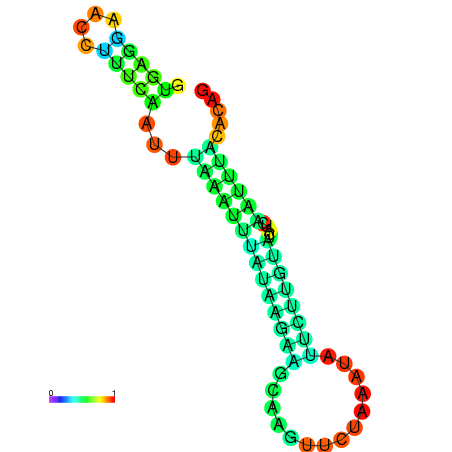
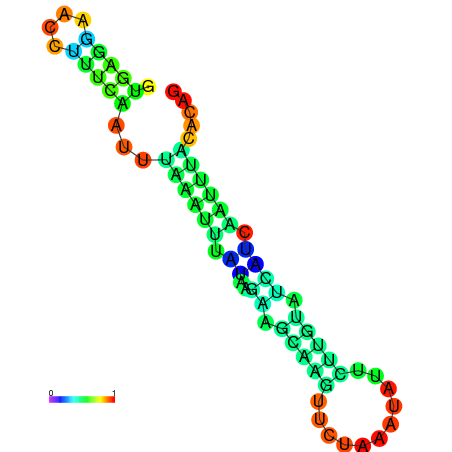
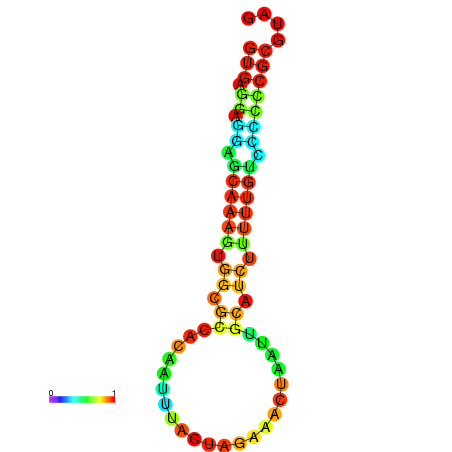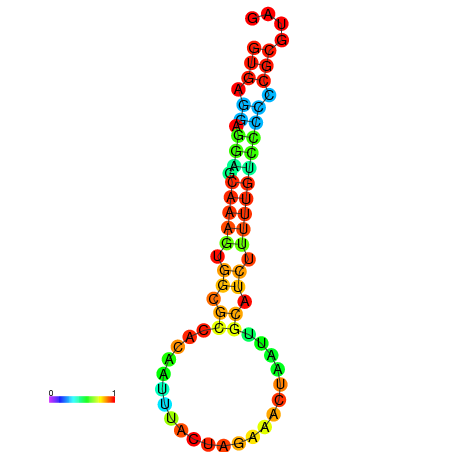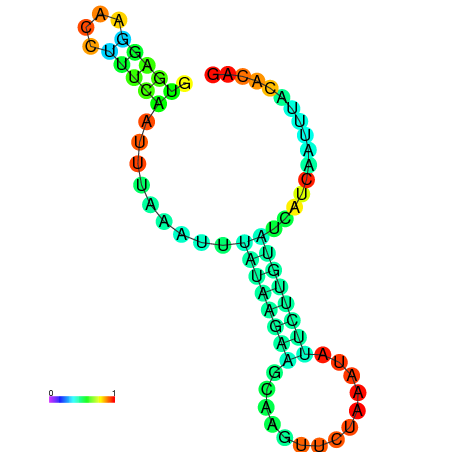| dm3 |
chr3R:11661994-11662091 - |
TGCGGATTCTGTCAGGTGAGGA---GGA-GCAAAGTGGCGCCACAA---------------------T-----------TTACTAGAAACTAATTGCA-----------TCTTTTTGTC------CCCCCGCGTAGGAAGCCTGTCCCGTG |
| droSim1 |
chr3R:9814135-9814231 + |
TGCGGATTCTGTCAGGTGAGGA---GGA-GCGCAGTGGCGCCACTA---------------------T-----------TTACTAGAAACTAATTGGA-----------TATTCTTGTC------CACCC-CGCAGGAAGCCTGTCCCGTG |
| droSec1 |
super_0:10292637-10292733 + |
TGCGGATTCTGTCAGGTGAGGA---GGA-GCGCAGTGGCGCCACTA---------------------T-----------TTACTAGATACTAATTGGA-----------TATTCTTGTC------CACCC-CGCAGGAAGCCTGTCCCGTG |
| droYak2 |
chr3R:14775389-14775483 + |
TGCGGATTCTGTCAGGTGAGGA---GGA-GCACACTCTTGCCACTG---------------------T-----------TTACGGGAAAATAATTGCA-----------TTTCCTTA--------CCCCC-TTCAGGAAGCCTGTCCCGTG |
| droEre2 |
scaffold_4770:11186817-11186911 - |
TGCGGATTCTGTCAGGTGAGGA---GAA-GCACAGTGGTGCCACTG---------------------T-----------GAAATGGAATCTTATAGCA-----------AATCCTT-T-------CCGCC-TTCAGGAAGCCTGTCCCGTG |
| droAna3 |
scaffold_13340:16266860-16266959 - |
TGCGGTTTCTGTCAGGTGAGGA---ACC-TTTCA---------------ATTTAAATTTATAAGAAGC-----------AAGTTC---------TAAA-----------TATTCTTGTATCATCAATTTA-CACAGGAGGCTTGCCCAGTT |
| dp4 |
chr2:20340730-20340823 - |
TGTGGCTTCTGTCAGGTAAGGAATGGGAGTCGAAATTTTTACATAA---------------------T-----------TTCAAGTCGATTCATTGAC-----------TT------T-------TTGCT-CGTAGGAGGCCTGCCCTGTG |
| droPer1 |
super_3:3092221-3092314 - |
TGTGGCTTCTGTCAGGTAAGGAATGGGAGTCGAAATTTTTACATCA---------------------T-----------TTCAAGTCGATTCATTGAC-----------TT------T-------TTGCT-CGCAGGAGGCCTGCCCTGTG |
| droWil1 |
scaffold_181130:15806459-15806551 + |
TGCGGATTCTGTCAGGTGAGTG---TGA-CTGAAATTCCAATTCAG---------------------C-----------ATATTTGAACTATATTA-------------T--------ATTTTCTGCCCC-TGCAGGAAGCTTGTCCCGTG |
| droVir3 |
scaffold_12855:5731320-5731404 + |
TGCGGTTTTTGTCAGGTAATTA---T------------------TA---------------------C-----------CCATAAGTATCCCATATAAAACTCATGGTGT------GTT------TCCTT-TGCAGGAGGCCTGTCCAGTC |
| droMoj3 |
scaffold_6540:6055055-6055141 + |
TGCGGTTTTTGTCAGGTAATT-----------------------TATCCATATGGATA-----------------TAATTTTGAGTGGGGT----T------CA-----TGATT--CTT------CCATT-AATAGGAGGCTTGCCCAGTC |
| droGri2 |
scaffold_14906:8272376-8272477 - |
TGCGGTTTTTGTCAGGTAATCT-----------------------ATTATTCTATATGTCCAGCAAGAAATTTAACAATTTTAATTATGTT----C-------------C--------ACATTTAAATGT-TGCAGGAGGCCTGTCCCGTC |