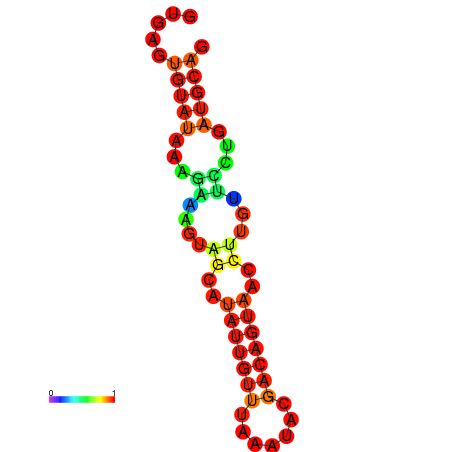
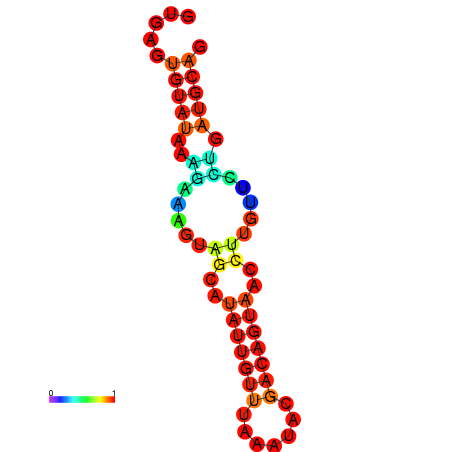
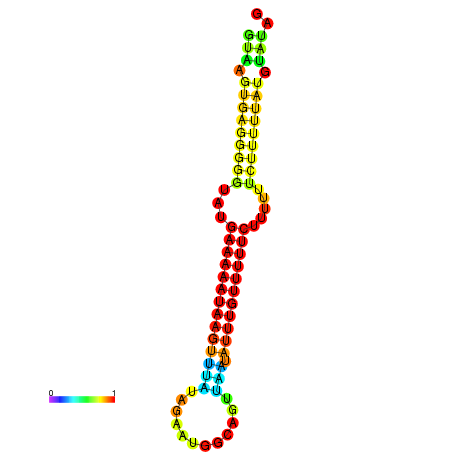
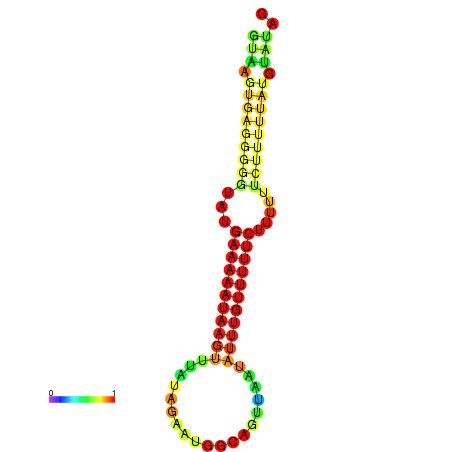
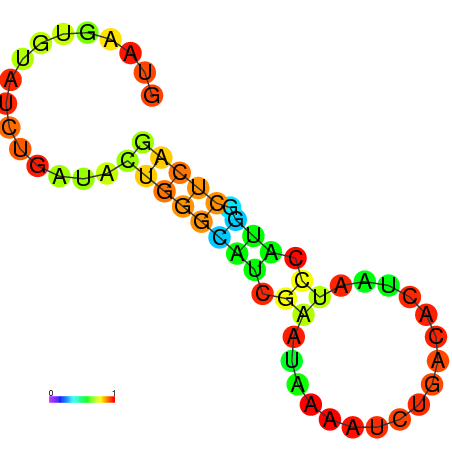
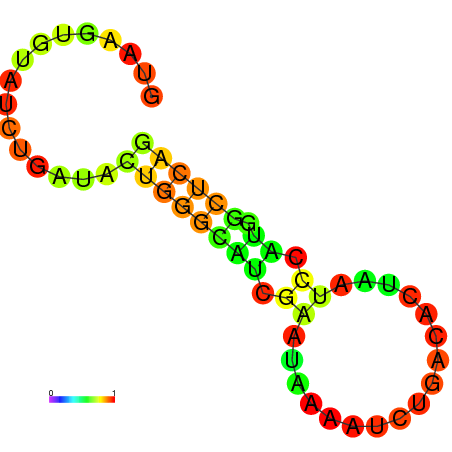
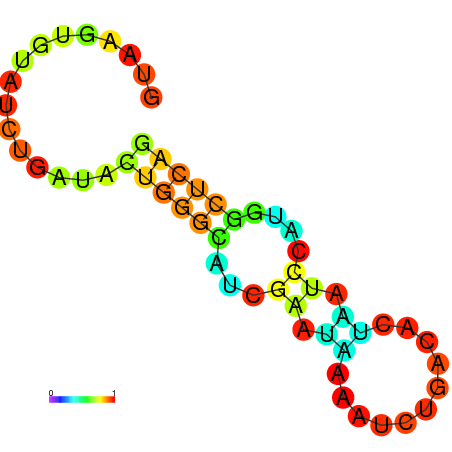
| dm3 |
chr2R:3598907-3598996 + |
GGCCATTAGGACAGGGTAGGTTGAA-----ATCGGATACAA---CTAAAAG-----TT-ATTTTTAATTGT-------------------AT--------CCG-GTCTT-C-ATTTTAGGTCTTCGATTGTTCC |
| droSim1 |
chr2R:2322894-2322983 + |
GGCCATTAGAACAGGGTAGGTAGAA-----ATCGGATTAAA---CTAAAAC-----TT-ATTTTTGATTGT-------------------CT--------CTT-GTTTT-C-ATTTGAGGTCTTCGATTGTGCC |
| droSec1 |
super_1:1254446-1254535 + |
GGCCATTAGAACAGGGTAGGTAGAA-----ATCGGATTAAA---CTAAAAC-----TT-ATTTTTGATTGT-------------------CT--------CTT-GTCTT-C-ATTTGAGGTCTTCGATTGTGCC |
| droYak2 |
chr2L:16271881-16271969 + |
GGCCATTAGGACAGGGTAGGTAGAT------TCGGGTTTAA---CTAAAAG-----TA-AGATTTAATAGT-------------------TT--------TCT-TTCCA-C-ACTTTAGGTCTTCAATTGTGCC |
| droEre2 |
scaffold_4929:19008519-19008607 - |
GGCCATTAGAACAGGGTAGGTAGGT------TCGGATATAA---CTAAAAG-----TA-AGATTTAATAGT-------------------TT--------TCT-TGCAT-C-ACTTTAGGTCTTCGATTGTGCC |
| droAna3 |
scaffold_13266:10106458-10106549 + |
GGCTATTCGAACGGGGTGAGTGAAAATTATTATGGA---AA---TTGATGG-----CA-------GAATCT-------------------CTCTAAAAAACT---CTTT-T-ATTGCAGTTCCTCAATTGTACC |
| dp4 |
chr3:6515146-6515237 - |
GGCCATAAGAACAGGGTGAGTGTAT-----AATGGAAGTAG---CACATTG--T--TT-AAATACGAAAGT-------------------AA--------CCTTGTTCC-T-GATGCAGTTCATCGATTGTGCC |
| droPer1 |
super_2:6735406-6735497 - |
GGCCATAAGAACAGGGTGAGTGTAT-----AAAGAAAGTAG---CATATTG--T--TT-AAATACGACAGT-------------------AA--------CCTTGTTCC-T-GATGCAGTTCATCGATTGTGCC |
| droWil1 |
scaffold_180700:1009012-1009118 + |
GGCTATACGTACGGGGTAAGTGAGG--------GGGTATGAAAAATAAGTTTATAG------AATGGCAGTTAATATTTGTTTT-----TCTTTT---TTCT---TTT--TATGTATAGTTCCTCTATAGTCCC |
| droVir3 |
scaffold_12875:823172-823263 + |
GGCCATACGCACGGGGTGAGTACGGCTG---ACTGCGATAA---CTGCACG-----CACATTCACGGCATT-------------------CA--------CTC--TCC--CATGAGCAGTTCGTCGATTGTGCC |
| droMoj3 |
scaffold_6496:3846141-3846250 + |
GGCCGTGCGAACAGGGTGAGTAGAC-----AGGAGA-AAAA---CTAACTGTATAACGCTTCCATTACATT-------CAACTATCCTCTCT--------CTTTGTCTCTTGTGAACAGTTCTGCTATTGTTCC |
| droGri2 |
scaffold_15245:10130930-10131015 - |
GGCCATACGCACGGGGTAAGTGTATCTGATACTGGGCATCG--------AA-----TA-AAATCTGACACT-------------------AA--------T-----CCA-T-GGCTCAGTTCGTCCATTGTGCC |