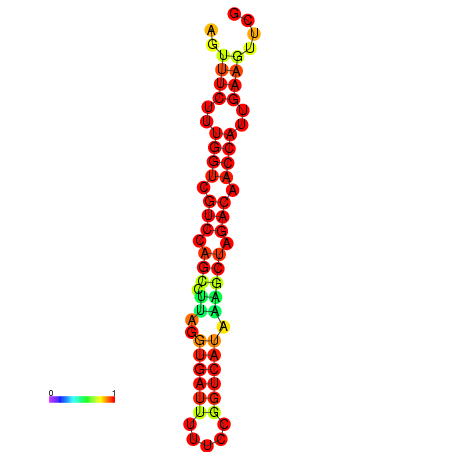
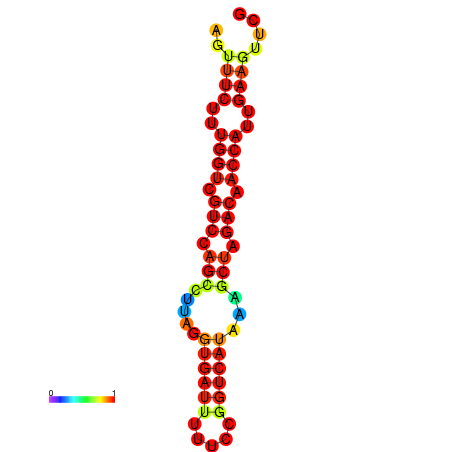
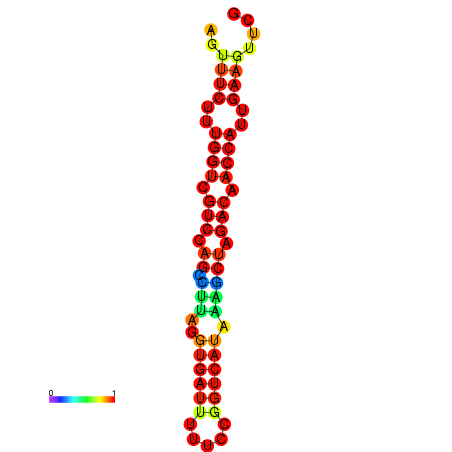
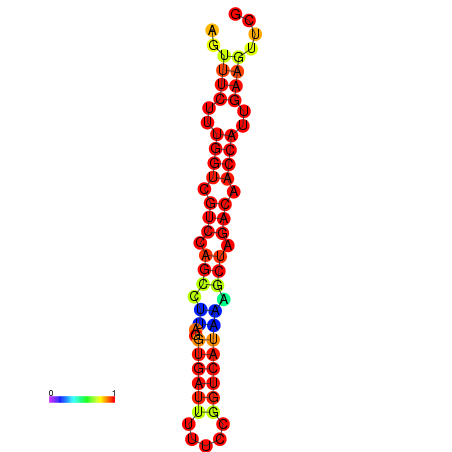
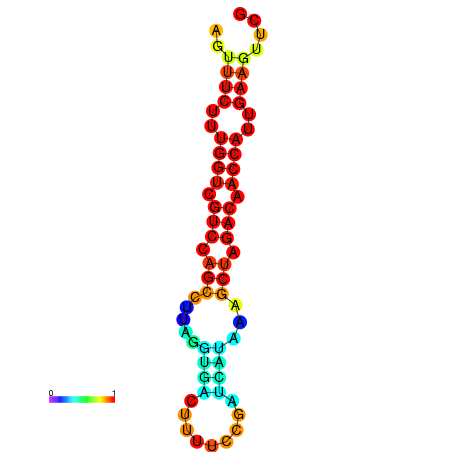
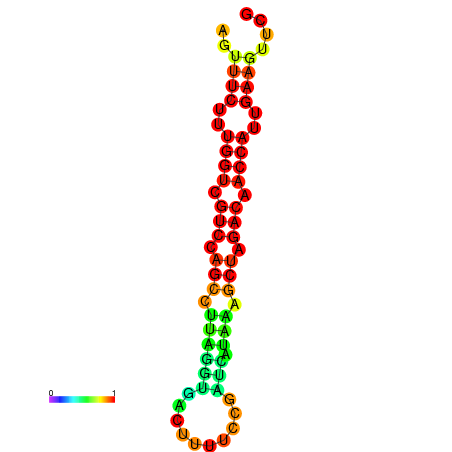
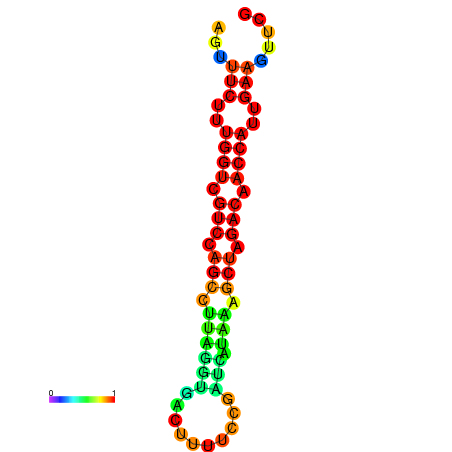
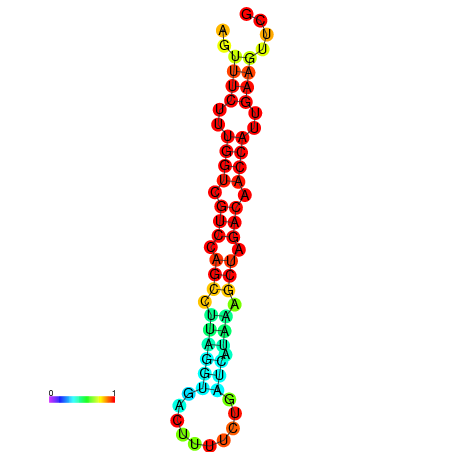
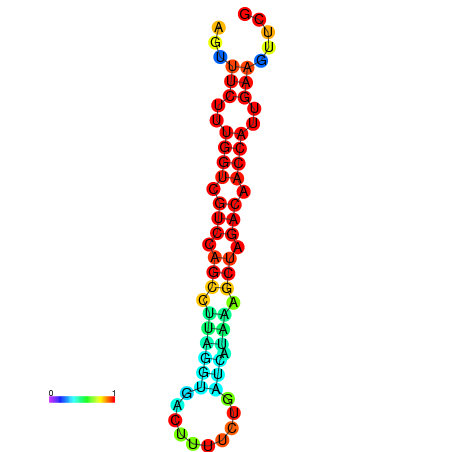
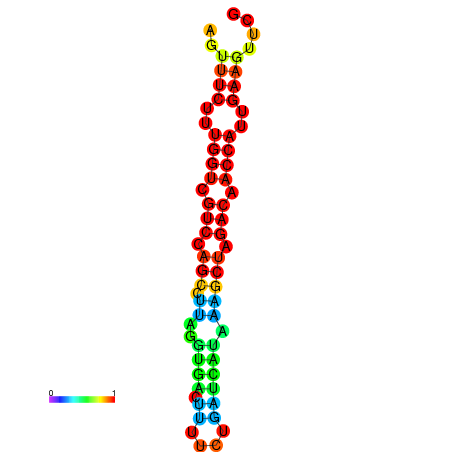
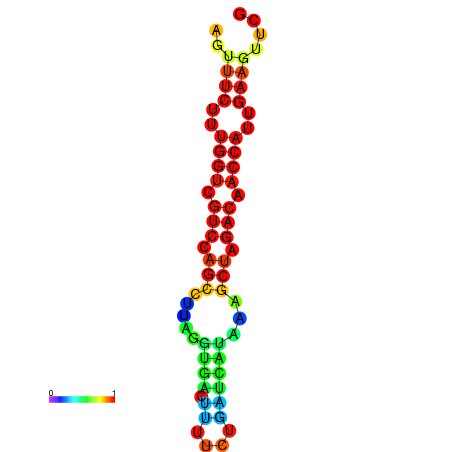
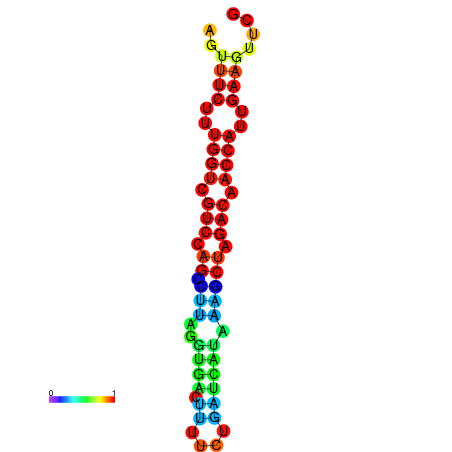
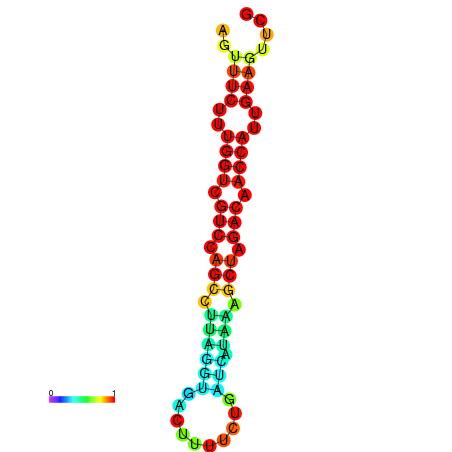
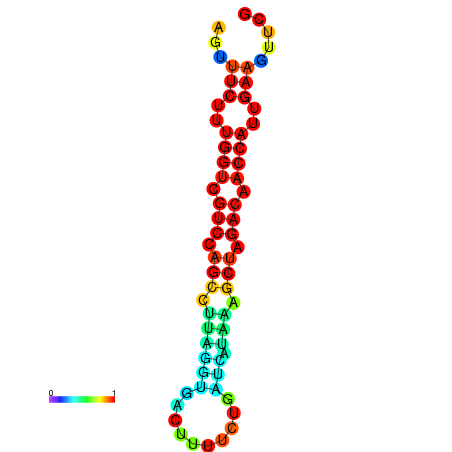
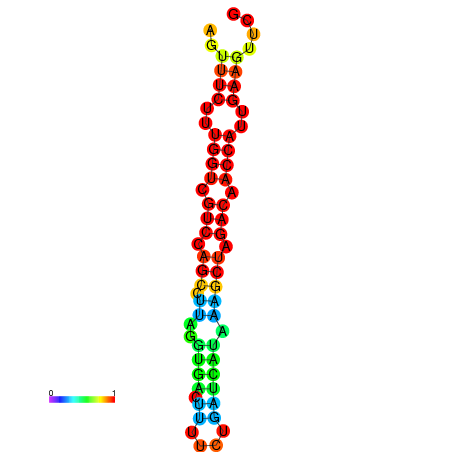
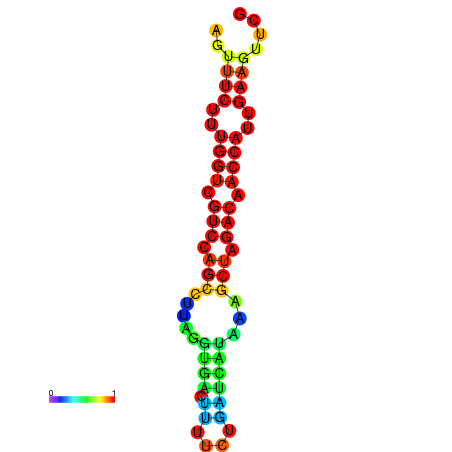
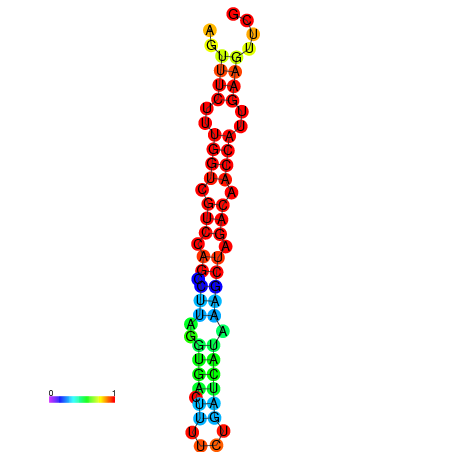
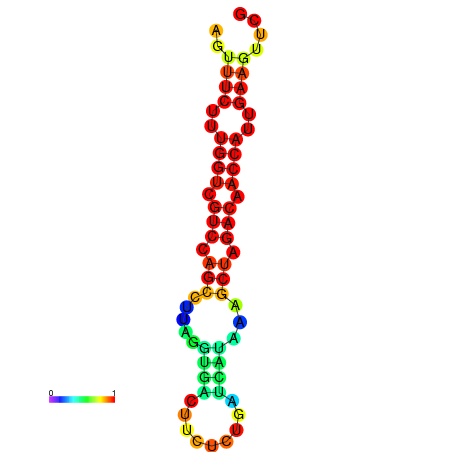
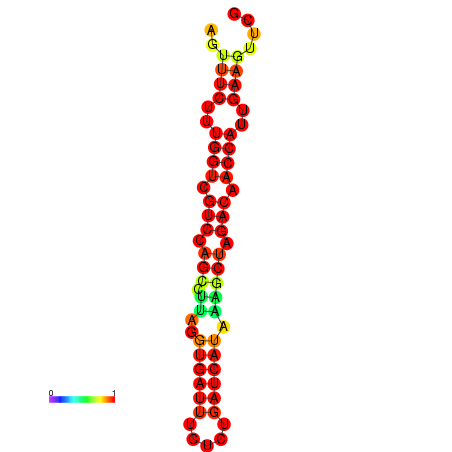
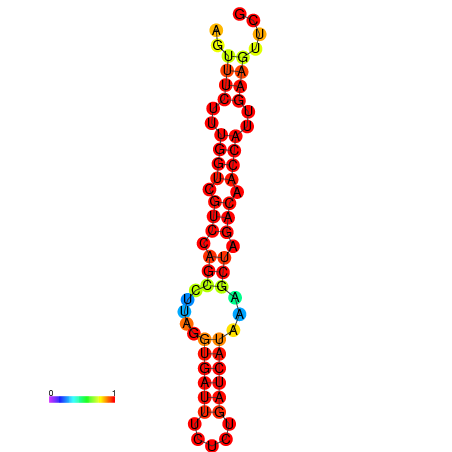
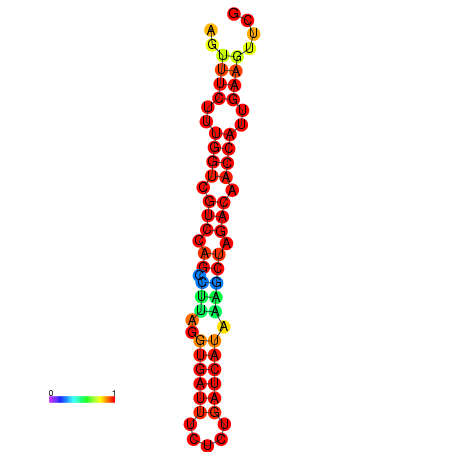
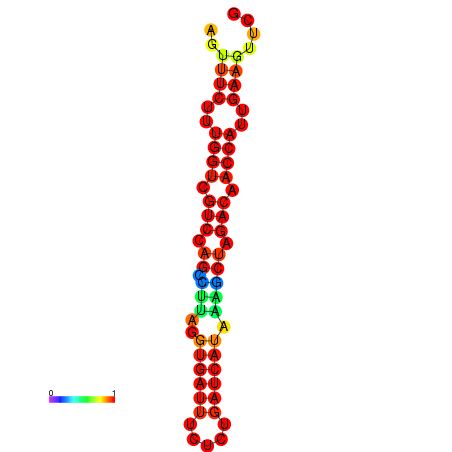
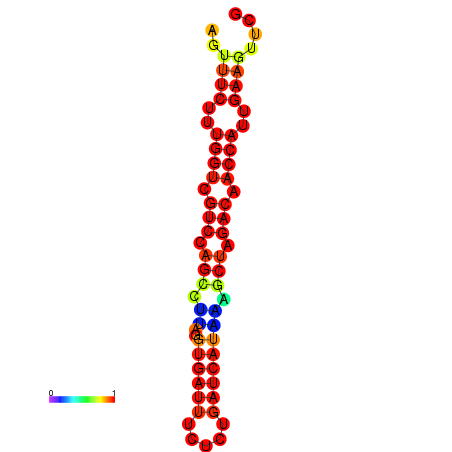
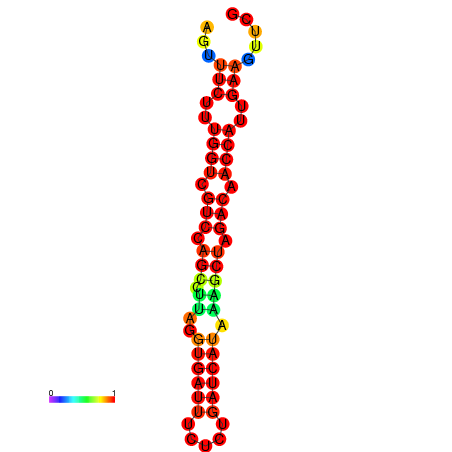
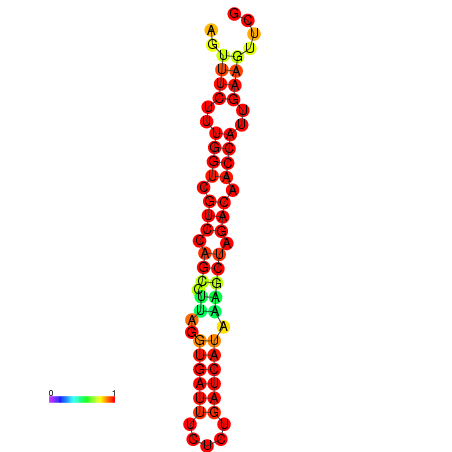
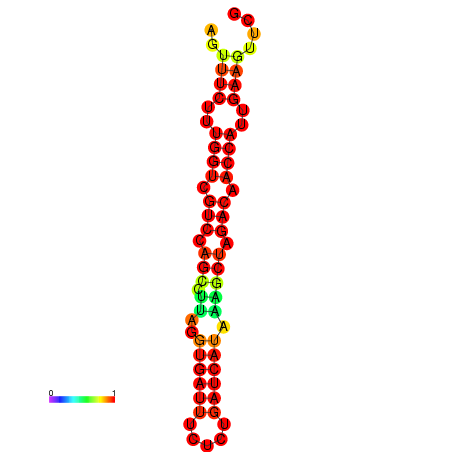
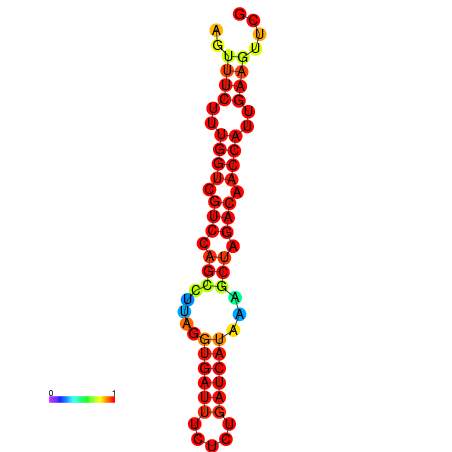
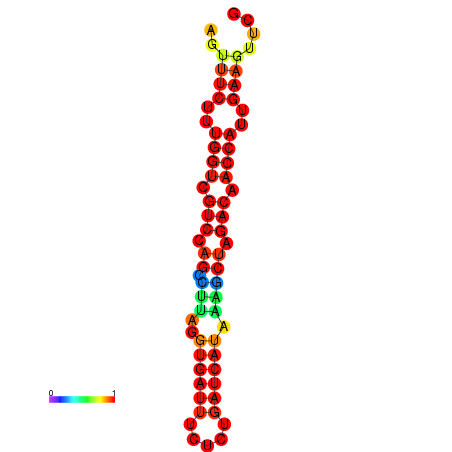
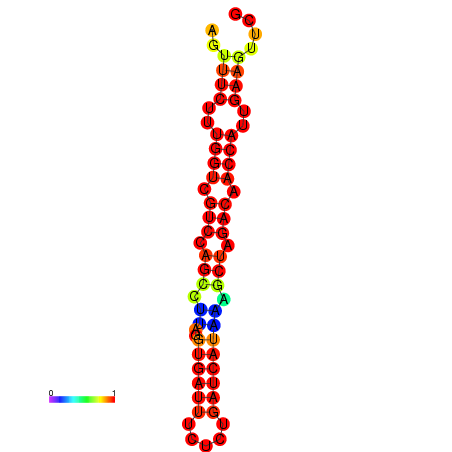
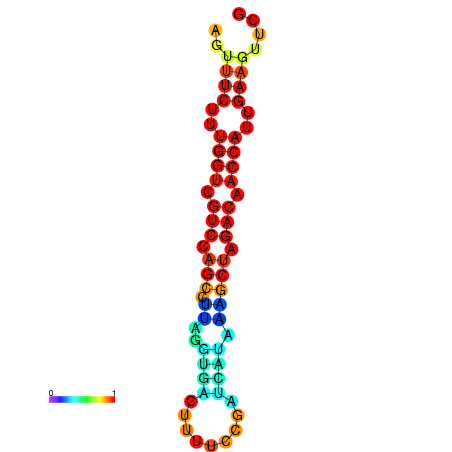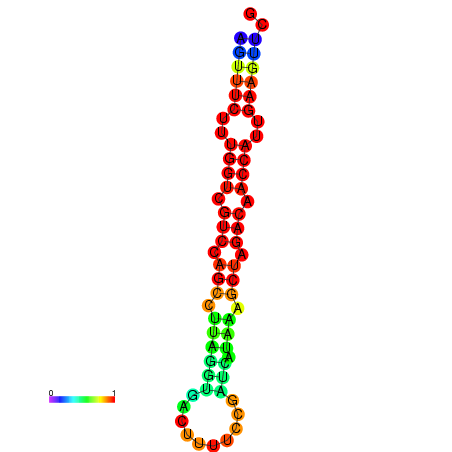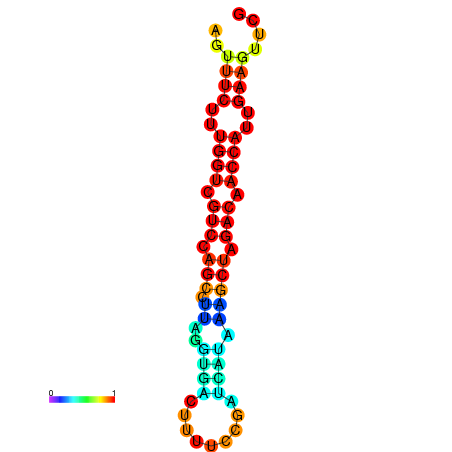| TACCTT--ATTGTGGTTGCAATTAGTTTCTTTGGTCGTCCAGCCTTAGGTGATTTCTCTGATCATAAAGCTAGACAACCATTGAAGTTCGTTGTGGCATTTGCAGCACTAC | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ...............................................................ATAAAGCTAGACAACCATTGAA.......................... | 22 | 1 | 3994.00 | 3994 | 3994 | 0 |
| ...............................................................ATAAAGCTAGACAACCATTGA........................... | 21 | 1 | 2063.00 | 2063 | 2063 | 0 |
| ...............................................................ATAAAGCTAGACAACCATTGAAG......................... | 23 | 1 | 636.00 | 636 | 636 | 0 |
| ................................................................TAAAGCTAGACAACCATTGAAG......................... | 22 | 1 | 227.00 | 227 | 227 | 0 |
| ...............................................................ATAAAGCTAGACAACCATTG............................ | 20 | 1 | 225.00 | 225 | 225 | 0 |
| ............................CTTTGGTCGTCCAGCCTTAGGTGA........................................................... | 24 | 1 | 222.00 | 222 | 222 | 0 |
| ................................................................TAAAGCTAGACAACCATTGAAGT........................ | 23 | 1 | 105.00 | 105 | 105 | 0 |
| ...............................................................ATAAAGCTAGACAACCATTGAAGT........................ | 24 | 1 | 66.00 | 66 | 66 | 0 |
| ...........................TCTTTGGTCGTCCAGCCTTAGGTGA........................................................... | 25 | 1 | 58.00 | 58 | 58 | 0 |
| ...........................TCTTTGGTCGTCCAGCCTTAGG.............................................................. | 22 | 1 | 32.00 | 32 | 32 | 0 |
| ............................CTTTGGTCGTCCAGCCTTAGGT............................................................. | 22 | 1 | 29.00 | 29 | 29 | 0 |
| ................................................................TAAAGCTAGACAACCATTGAA.......................... | 21 | 1 | 24.00 | 24 | 24 | 0 |
| ...........................TCTTTGGTCGTCCAGCCTTAG............................................................... | 21 | 1 | 24.00 | 24 | 24 | 0 |
| ............................CTTTGGTCGTCCAGCCTTAGGTG............................................................ | 23 | 1 | 21.00 | 21 | 21 | 0 |
| ...........................TCTTTGGTCGTCCAGCCTTAGGT............................................................. | 23 | 1 | 14.00 | 14 | 14 | 0 |
| ...........................TCTTTGGTCGTCCAGCCTTAGGTG............................................................ | 24 | 1 | 8.00 | 8 | 8 | 0 |
| ................................................................TAAAGCTAGACAACCATTGA........................... | 20 | 1 | 8.00 | 8 | 8 | 0 |
| ...............................................................ATAAAGCTAGACAACCATT............................. | 19 | 1 | 7.00 | 7 | 7 | 0 |
| ............................CTTTGGTCGTCCAGCCTTAGG.............................................................. | 21 | 1 | 6.00 | 6 | 6 | 0 |
| .............................TTTGGTCGTCCAGCCTTAGGTGA........................................................... | 23 | 1 | 6.00 | 6 | 6 | 0 |
| .........TTGTGGTTGCAATTAGTTT................................................................................... | 19 | 1 | 5.00 | 5 | 5 | 0 |
| ..............................................................CATAAAGCTAGACAACCATTGA........................... | 22 | 1 | 4.00 | 4 | 4 | 0 |
| .................................................................AAAGCTAGACAACCATTGAAGT........................ | 22 | 1 | 4.00 | 4 | 4 | 0 |
| ..................................................................AAGCTAGACAACCATTGAAGT........................ | 21 | 1 | 4.00 | 4 | 4 | 0 |
| ..............................................................CATAAAGCTAGACAACCATTGAA.......................... | 23 | 1 | 3.00 | 3 | 3 | 0 |
| ............................CTTTGGTCGTCCAGCCTTAG............................................................... | 20 | 1 | 3.00 | 3 | 3 | 0 |
| ..................................................................AAGCTAGACAACCATTGAA.......................... | 19 | 1 | 2.00 | 2 | 2 | 0 |
| ..........TGTGGTTGCAATTAGTTT................................................................................... | 18 | 1 | 2.00 | 2 | 2 | 0 |
| .....T--ATTGTGGTTGCAATTAGTTT................................................................................... | 21 | 1 | 2.00 | 2 | 2 | 0 |
| ..................................................................AAGCTAGACAACCATTGA........................... | 18 | 1 | 1.00 | 1 | 1 | 0 |