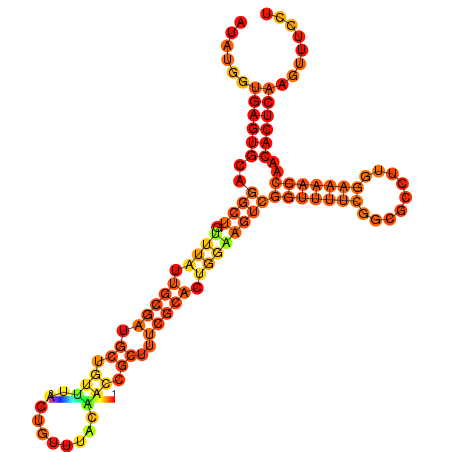
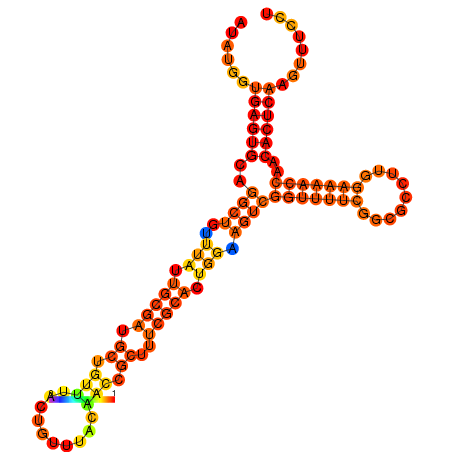
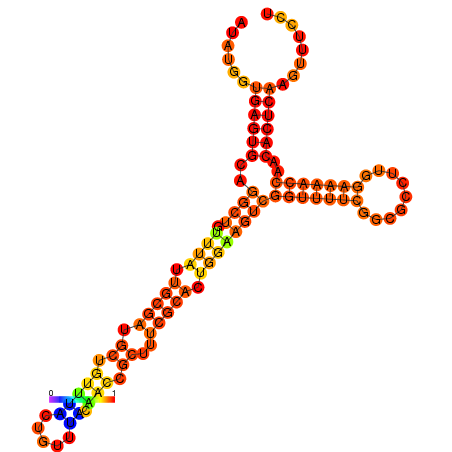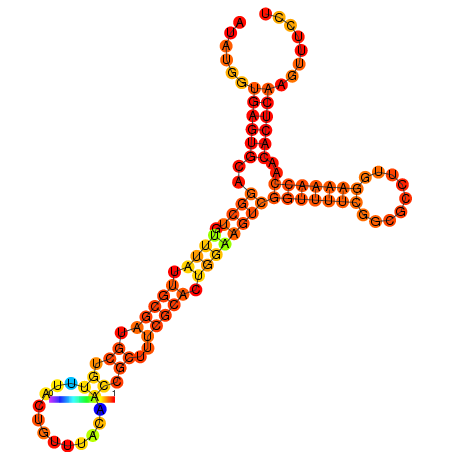| dm3 |
chrX:5516298-5516464 - |
AAACAAATCGATATGGTGAGTGC---AG-GC----------------TGTTTAT----------TGCGGCGCTGTTTACTGTTTACGATC------GCACTGGAAGTGCGCTTTCCGGCGCCTTTGGAAGCCAG-----------------------------------------CACTCAAGTTTCCTTCATTGTTTTCCTTTCTTTCCCCTCTCTCTCTC------C---CCGCAGGCAGCTTATACCGAG |
| droSim1 |
chrX:4275639-4275796 - |
AAACAAATAGAAATGGTGAGTGC---AG-GC----------------TGTTTAT----------TGCGGCGCTGTTTACTGTTTACGATC------G---------TGCGCTATTCGGCGCCTTTGGAAGCCAG-----------------------------------------CACTCAAGTTTCCCTCATTGTTTTCCTTTCTTCCCCCCCTCTCTCTC------C---CCGCAGGCAGCTTACACCGAG |
| droSec1 |
super_4:1240682-1240837 + |
AAACAAATAGAAATGGTGAGTGC---AG-GC----------------TGTTTAT----------TGCGGCGCTGTTTACTGTTTACGATC------G---------TGCGCTATTCGGCGCCTTTGGAAGCCAG-----------------------------------------CACTCAAGTTTCCCTCATTGTTTTCCTTTCTTTCC-CCCTCTCTCA-------C---CCGCAGGCAGCATACACCGAG |
| droYak2 |
chrX:2953226-2953406 + |
AAACAAATCGATATGGTGAGTGC---AG-GC----------------TGTTTAT----------TGCGATGCTGTTTACTGTTTACAACCGCTTTCGCACTGGAAGT-CGGTTTTCGGCGCCTTGGAAAACCAA-----------------------------------------CACTCAAGTTTCCTTCTTTGTTTTCCTTTTCTCTCCCTCTCTCTCTTTCTCTCTCTGTCGCAGGCCGCTTACACCGAG |
| droEre2 |
scaffold_4690:2867616-2867787 - |
AAACAAATCGATATGGTGAGTGC---AG-GC----------------TGTTTAT----------TGCGGCGCTGTTTACTGTTTACGGTC------GCACCGCAAGT-CGCTTTTCGGCGCCTTGGAAAACCAA-----------------------------------------CACTCAAGTTTCCCTCTTTGTTTTCCTTTCTTCTCCCTCTCTCTCTCACTCTCT---TCGCAGGCAGCTTACACCGAG |
| droAna3 |
scaffold_13117:2022234-2022400 + |
AAACAAATCGATATGGTAAGTGCGTGAA-GTAGGCAACATGACACTTTGTATAT----------TTGACAACAATTGAATGTTTACGGG--------------ACCTC--CTATTTCCCGC-TTCGGCAGCAACGA-TG-----------ACGACGAC-----------------GACTCATGTTTTCCT--TTGTTTTGTTGTT---------------------------TTTCAGGCAGCTTACACCGAG |
| dp4 |
chrXL_group1a:8129365-8129509 - |
CAACAAATCGATATGGTAAGTGGGCTGA-AC----------------AGTA-AT------------TGATATCGTGGCCAATATATAGCCGCT-ACGTATCGTATG---------------------------------------------GCTTTTCCACTGAGTTTTGCTGGGAGCTAATGTGTTCTT--TTGTT--TGG-T------------------------T---TTACAGGCAGCTTACACCGAG |
| droPer1 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_12928:4987995-4988105 + |
T---CAATCAATATGGTAAGTTGGCCAAAAT----------------TGTT-GTTGCT------TTT------------ACGACACGTACATT-GTTTAT--------------------------AGTGTGTTTATTCTTGCTACTTT--------------------------------------------------TGC-A------------------------T---TTACAGGCAGCTTACACCGAG |
| droMoj3 |
scaffold_6359:3485302-3485418 + |
-------TCAATATGGTAAGTTGGC-ACAGC----------------TGTT----------------------------------------------------------GCTATT-----TTTTAGAGAGCAAATA-ATATGTTTCGTTTGTTTTTACAATTG----------------------------------------TTGTCTTTTTCTCTTGTTA------T---TTACAGGCAGCATACACCGAG |
| droGri2 |
scaffold_14853:9687493-9687620 + |
CATTAAATCAACATGGTAAGTTGTATAA-AT----------------CTATTTTTGCAGCTATCTGCAGCATTGTGTGCACTATATACACATT-AAT----------------------------GAGTGTAATAA-TTTTGTTGTTTT--------------------------------------------------TGT-T------------------------T---TTACAGGCAGCTTACACCGAG |