
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:5926643-5926771 + | CAAACAA-GTATTC------TTAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TT--ATTGC-CGTTGACAA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAAAGCAA----------CTTATAA--- |
| droSim1 | chr3R:15449371-15449500 - | CAATCAAAGTATTC------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TT--ATTGC-CGTTGACAA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAAAGCAA----------CTTATGG--- |
| droSec1 | super_0:15986833-15986962 - | CAATCAAAGTATTC------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TT--ATTGC-CGTTGACAA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAAAGCAA----------CTTATAG--- |
| droYak2 | chr3R:9974454-9974582 + | CAATCAA-GTATTC------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TT--ATTGC-CGTTGACAA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAAAGCAA----------CGTACTC--- |
| droEre2 | scaffold_4770:15673299-15673427 - | CAATCAA-GTATTC------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TC--ATTGC-TTGTGACAA----CTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAAAGCAA----------CGTACTC--- |
| droAna3 | scaffold_13340:6489967-6490093 + | TAATTCA-GTTACC------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCTGG-------TGCTCGTTGGCAATTAATTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAGAGCAA----------CTTA------ |
| dp4 | chr2:5019555-5019683 - | AAATCCA-GAAAAT------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAAAAT--ATTGC-CTTTGACCA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCCATAACAA----------CG-ATAA--- |
| droPer1 | super_19:724982-725124 - | AAATCCA-GAAAAT------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAAAAT--ATTGC-CTTTGACCA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCCATAACAACGATAAGCAACATATAATCG |
| droWil1 | scaffold_181089:11166523-11166648 - | TAAC------AT--------TCTATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TTAAATTGC-CAA-AACAA-TAAACCACAGCCACTATCTTCACTGCCGCCGCGACAGGCCATGACAA----------CAATCGG--- |
| droVir3 | scaffold_12855:2682541-2682664 - | AAAAAAA-ATTACCTTAAATCCGATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTGGCCAA-T------------TGACAA----ATCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTATGACAG----------CAAA--A--- |
| droMoj3 | scaffold_6540:11883356-11883481 - | CGAAAAA-ATACCCTTTAATCCGATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTGGCCAC-T------------TGAAAA----ATCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTATGACAC----------CGAGAGA--- |
| droGri2 | scaffold_14624:2267430-2267555 + | AAACCAA-ATTCCAT-----TGAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTGGCCAA-C------------TGACAA----ATCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTATGACACCAATAA-----ATATAA--- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:5926643-5926771 + | CAAACAA-GTATTC------TTAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TT--ATTGC-CGTTGACAA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAAAGCAA----------CTTATAA--- |
| droSim1 | chr3R:15449371-15449500 - | CAATCAAAGTATTC------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TT--ATTGC-CGTTGACAA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAAAGCAA----------CTTATGG--- |
| droSec1 | super_0:15986833-15986962 - | CAATCAAAGTATTC------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TT--ATTGC-CGTTGACAA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAAAGCAA----------CTTATAG--- |
| droYak2 | chr3R:9974454-9974582 + | CAATCAA-GTATTC------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TT--ATTGC-CGTTGACAA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAAAGCAA----------CGTACTC--- |
| droEre2 | scaffold_4770:15673299-15673427 - | CAATCAA-GTATTC------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TC--ATTGC-TTGTGACAA----CTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAAAGCAA----------CGTACTC--- |
| droAna3 | scaffold_13340:6489967-6490093 + | TAATTCA-GTTACC------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCTGG-------TGCTCGTTGGCAATTAATTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTAGAGCAA----------CTTA------ |
| dp4 | chr2:5019555-5019683 - | AAATCCA-GAAAAT------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAAAAT--ATTGC-CTTTGACCA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCCATAACAA----------CG-ATAA--- |
| droPer1 | super_19:724982-725124 - | AAATCCA-GAAAAT------TCAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAAAAT--ATTGC-CTTTGACCA----TTCACAGCCACTATCTTCACTGCCGCCGCGACAAGCCATAACAACGATAAGCAACATATAATCG |
| droWil1 | scaffold_181089:11166523-11166648 - | TAAC------AT--------TCTATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTAGCCAA-TTAAATTGC-CAA-AACAA-TAAACCACAGCCACTATCTTCACTGCCGCCGCGACAGGCCATGACAA----------CAATCGG--- |
| droVir3 | scaffold_12855:2682541-2682664 - | AAAAAAA-ATTACCTTAAATCCGATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTGGCCAA-T------------TGACAA----ATCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTATGACAG----------CAAA--A--- |
| droMoj3 | scaffold_6540:11883356-11883481 - | CGAAAAA-ATACCCTTTAATCCGATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTGGCCAC-T------------TGAAAA----ATCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTATGACAC----------CGAGAGA--- |
| droGri2 | scaffold_14624:2267430-2267555 + | AAACCAA-ATTCCAT-----TGAATTGGCTATGCGCTTTGGCAGTGTGGTTAGCTGGTTGTGTGGCCAA-C------------TGACAA----ATCACAGCCACTATCTTCACTGCCGCCGCGACAAGCTATGACACCAATAA-----ATATAA--- |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-34.4, p-value=0.009901 | dG=-33.4, p-value=0.009901 |
|---|---|
 |
 |
| dG=-34.4, p-value=0.009901 | dG=-33.4, p-value=0.009901 |
|---|---|
 |
 |
| dG=-34.4, p-value=0.009901 | dG=-33.4, p-value=0.009901 |
|---|---|
 |
 |
| dG=-34.4, p-value=0.009901 | dG=-33.4, p-value=0.009901 |
|---|---|
 |
 |
| dG=-37.4, p-value=0.009901 | dG=-36.4, p-value=0.009901 |
|---|---|
 |
 |
| dG=-38.8, p-value=0.009901 | dG=-37.9, p-value=0.009901 | dG=-37.8, p-value=0.009901 |
|---|---|---|
 |
 |
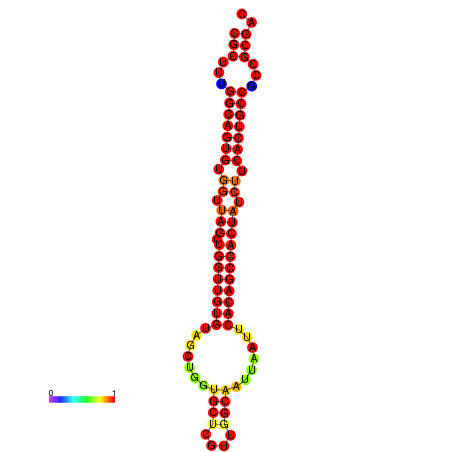 |
| dG=-34.8, p-value=0.009901 | dG=-33.8, p-value=0.009901 |
|---|---|
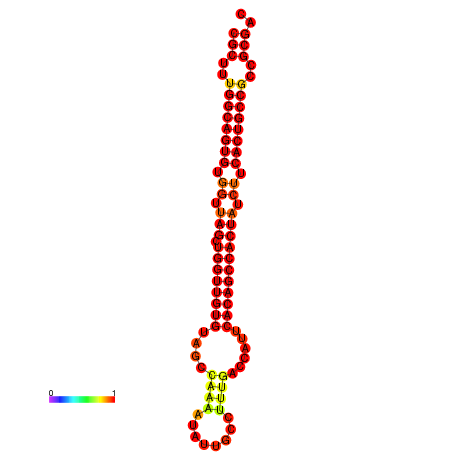 |
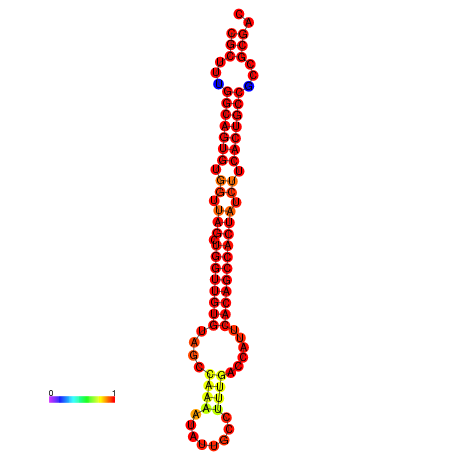 |
| dG=-34.8, p-value=0.009901 | dG=-33.8, p-value=0.009901 |
|---|---|
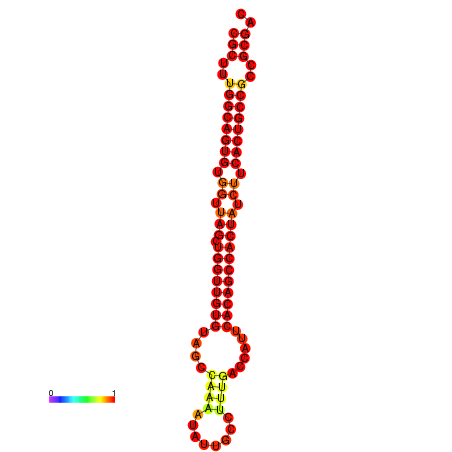 |
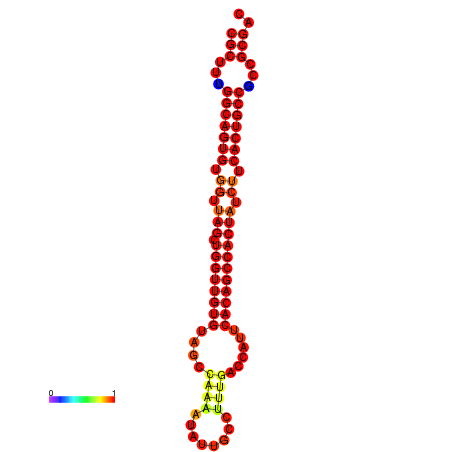 |
| dG=-32.4, p-value=0.009901 | dG=-32.2, p-value=0.009901 | dG=-31.4, p-value=0.009901 |
|---|---|---|
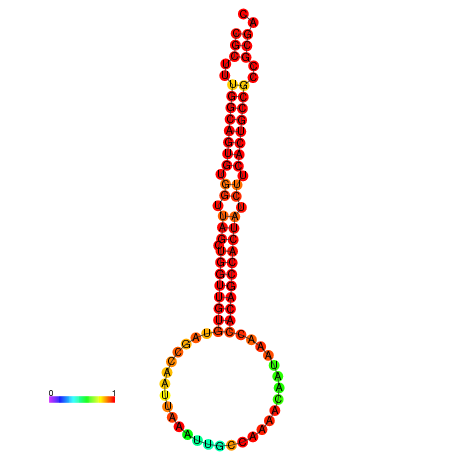 |
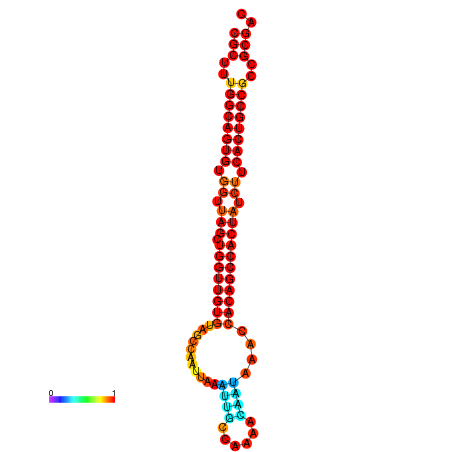 |
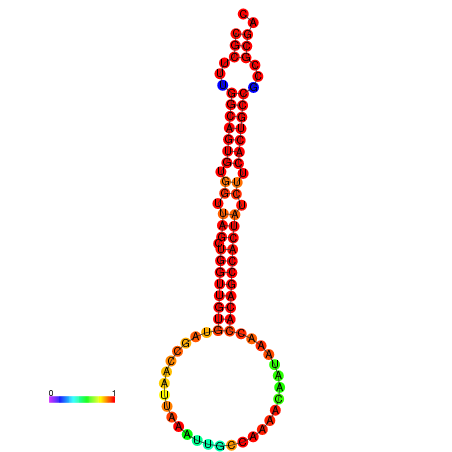 |
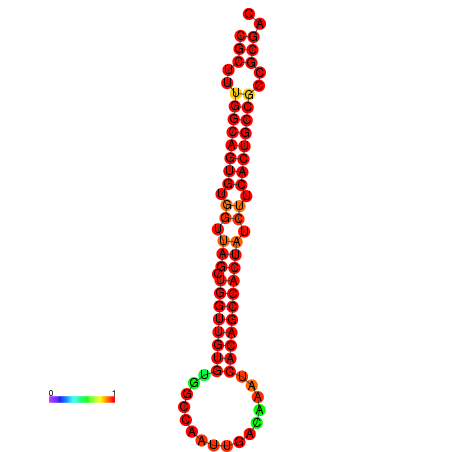
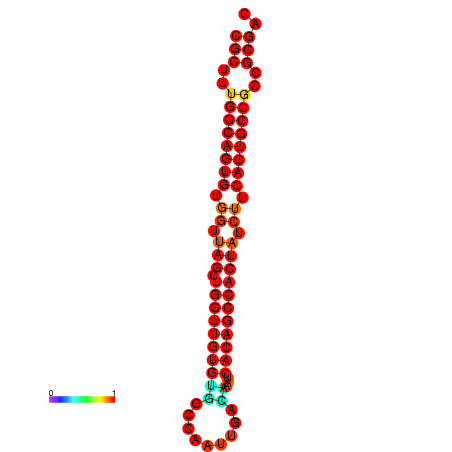
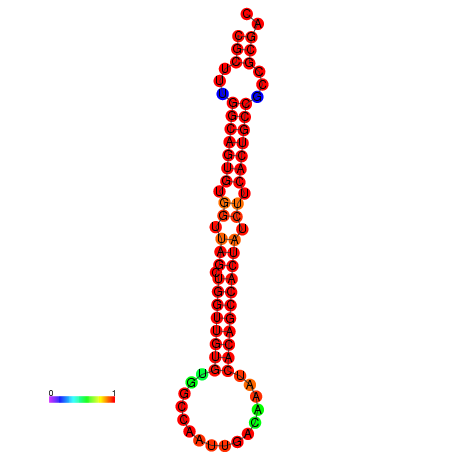
| dG=-33.6, p-value=0.009901 | dG=-33.5, p-value=0.009901 | dG=-32.6, p-value=0.009901 |
|---|---|---|
 |
 |
 |
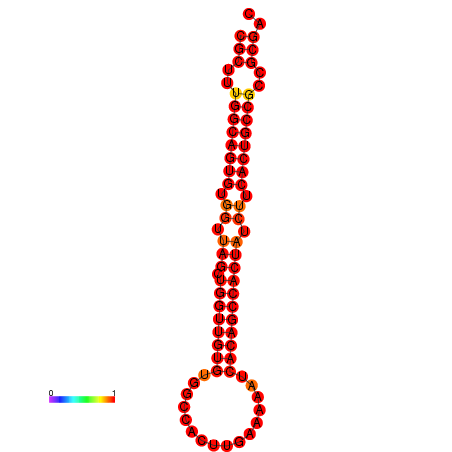
| dG=-33.6, p-value=0.009901 | dG=-32.6, p-value=0.009901 |
|---|---|
 |
 |
| dG=-33.6, p-value=0.009901 | dG=-33.5, p-value=0.009901 | dG=-32.6, p-value=0.009901 |
|---|---|---|
 |
 |
 |
Generated: 03/07/2013 at 05:22 PM