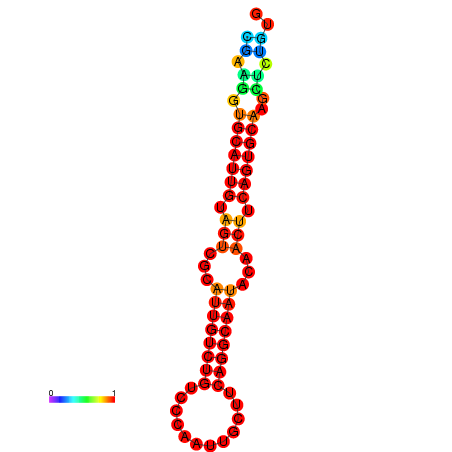
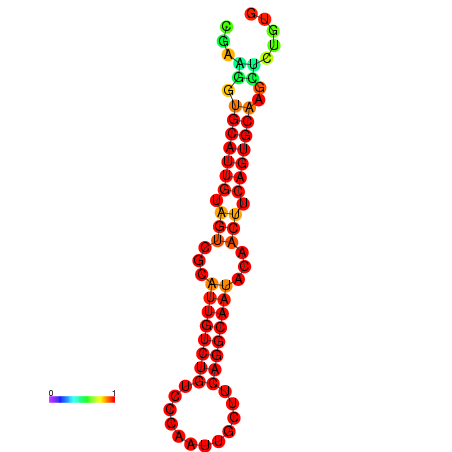
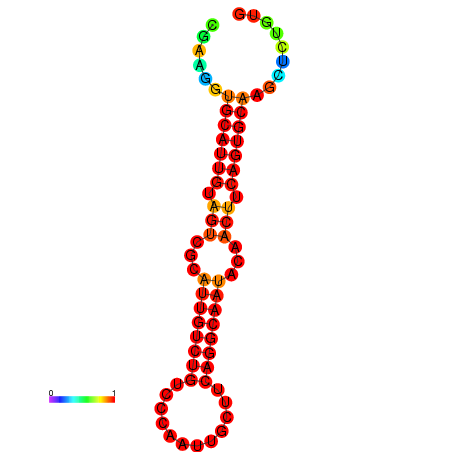
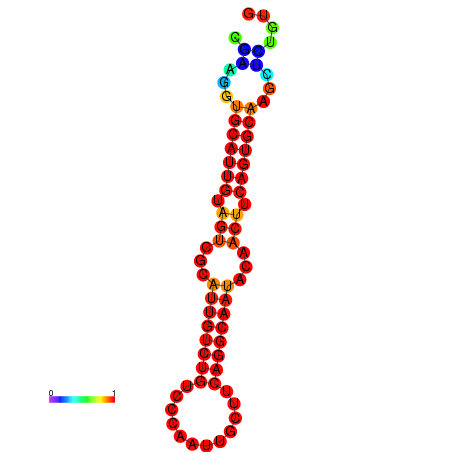
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:19791046-19791174 + | CTG-----AGTGCCACCAGACTCT--------TCCT-CTGGAGATGACACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCAATT---GC-TTCAGGCAATACAACTTCAGTGCAAGCTCTGTGCATTTCACATA-CAAC------------GCCG---AATG------ |
| droSim1 | chr3L:19083102-19083230 + | CTG-----AGTGCCACCAGACTCC--------TCCT-CCGGAGATGACACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCAATT---GC-TTCAGGCAATACAACTTCAGTGCAAGCTCTGTGCATTTCACACA-CAAC------------GCCG---AATG------ |
| droSec1 | super_29:452742-452870 + | CTG-----AGTGCCACCAGACTCT--------TCCT-CCGGAGATGACACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCAATT---GC-TTCAGGCAATACAACTTCAGTGCAAGCTCTGTGCATTTCACACA-CAAC------------GCCG---AATG------ |
| droYak2 | chr3L:20771315-20771443 - | CTG-----AGTGCCACCAGACTCT--------TACT-CCAGAGATGTCACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCAATT---GC-TTCAGGCAATACAACTTCAGTGCAAGCTCTGTGCATTTCACAAA-CAAC------------GCCG---AGTG------ |
| droEre2 | scaffold_4784:19550563-19550691 + | CTG-----AGTGCCACCAGACTCT--------CACT-CCAGAGATGACACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCAATT---GC-TTCAGGCAATACAACTTCAGTGCAAGCTCTGTGCATTTCACAAA-CAAC------------GCCG---AGTG------ |
| droAna3 | scaffold_13337:19456783-19456919 - | CCG-----AATACCAGCAGACACTATACACCCGCCC-TCGGAGATGTCACGCAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCACTT---GC-TCCAGGCAATACAACTGCAGTGCAAGCTCTGTGCATTTCACAAA-CAAC------------GCCA---ACTT------ |
| dp4 | chrXR_group6:5913809-5913939 + | AAA-----AACCAAACCAAACACT-----------C-CCGGAGATGACACGCAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CATCTT---GATTTCAGGCAATACAACTGCAATGCAAGCTCTGTGCATTTCATAAATCAAC------------GCCGTCCAATG------ |
| droPer1 | super_47:413986-414116 + | AAA-----AACCAAACCAAACACT-----------C-CCGGAGATGACACGCAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CATCTT---GATTTCAGGCAATACAACTGCAGTGCAAGCTCTGTGCATTTCATAAATCAAC------------GCCGTCCAATG------ |
| droWil1 | scaffold_180698:8572488-8572628 + | CGG-----AACCTAAACAAAT------------------TGAAGTTACACGAAGGTGCATTGTAGTCGCATTGTCTGTGCTCCAAAAAAAAAAAACCCCCAGTT-T-CTATTCAGGCAATACAACTGCAGTGCAAGCTTCGTGTGACTTATAAG-CAAC------------GGCA---AATA------ |
| droVir3 | scaffold_13049:22941104-22941227 - | CCA-----AAATC----CGACA-----------TCT-CTGGAGCTGTTACGGAGGTGCATTGTAGTCGCATTGTCTGTC-------------------TCAAAT-TTGC-CCGCGGCAATACAACTGCAGTGCAAGCTCCGTGCAGTTCACACG-CAAC------------GTCG---GAAG------ |
| droMoj3 | scaffold_6680:17552824-17552948 - | CAG-----CATTCCGACA---------------TCT-TTGGAGCTGTGACGGAGGTGCATTGTAGTCGCATTGTCTGTA-------------------TCCTTTATTCT-CTGCGGCAATACAACTGCAGTGCAAGCTCCGTGCAGTTCACTCG-CAAC------------GTCG---GACA------ |
| droGri2 | scaffold_15110:18214439-18214590 - | CTCTGCACAGTTAGGGCAGACA-----------TCTCCAGGAGCTGTCACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CAAAAA-TTGC-ATGCGGCAATACAACTGCAGTGCAAGCTCCGTGCAGTTCACTAG-CAACAACTCTTTGATCGCTG---ATCGATCTGA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:19791046-19791174 + | CTG-----AGTGCCACCAGACTCT--------TCCT-CTGGAGATGACACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCAATT---GC-TTCAGGCAATACAACTTCAGTGCAAGCTCTGTGCATTTCACATA-CAAC------------GCCG---AATG------ |
| droSim1 | chr3L:19083102-19083230 + | CTG-----AGTGCCACCAGACTCC--------TCCT-CCGGAGATGACACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCAATT---GC-TTCAGGCAATACAACTTCAGTGCAAGCTCTGTGCATTTCACACA-CAAC------------GCCG---AATG------ |
| droSec1 | super_29:452742-452870 + | CTG-----AGTGCCACCAGACTCT--------TCCT-CCGGAGATGACACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCAATT---GC-TTCAGGCAATACAACTTCAGTGCAAGCTCTGTGCATTTCACACA-CAAC------------GCCG---AATG------ |
| droYak2 | chr3L:20771315-20771443 - | CTG-----AGTGCCACCAGACTCT--------TACT-CCAGAGATGTCACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCAATT---GC-TTCAGGCAATACAACTTCAGTGCAAGCTCTGTGCATTTCACAAA-CAAC------------GCCG---AGTG------ |
| droEre2 | scaffold_4784:19550563-19550691 + | CTG-----AGTGCCACCAGACTCT--------CACT-CCAGAGATGACACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCAATT---GC-TTCAGGCAATACAACTTCAGTGCAAGCTCTGTGCATTTCACAAA-CAAC------------GCCG---AGTG------ |
| droAna3 | scaffold_13337:19456783-19456919 - | CCG-----AATACCAGCAGACACTATACACCCGCCC-TCGGAGATGTCACGCAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CCACTT---GC-TCCAGGCAATACAACTGCAGTGCAAGCTCTGTGCATTTCACAAA-CAAC------------GCCA---ACTT------ |
| dp4 | chrXR_group6:5913809-5913939 + | AAA-----AACCAAACCAAACACT-----------C-CCGGAGATGACACGCAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CATCTT---GATTTCAGGCAATACAACTGCAATGCAAGCTCTGTGCATTTCATAAATCAAC------------GCCGTCCAATG------ |
| droPer1 | super_47:413986-414116 + | AAA-----AACCAAACCAAACACT-----------C-CCGGAGATGACACGCAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CATCTT---GATTTCAGGCAATACAACTGCAGTGCAAGCTCTGTGCATTTCATAAATCAAC------------GCCGTCCAATG------ |
| droWil1 | scaffold_180698:8572488-8572628 + | CGG-----AACCTAAACAAAT------------------TGAAGTTACACGAAGGTGCATTGTAGTCGCATTGTCTGTGCTCCAAAAAAAAAAAACCCCCAGTT-T-CTATTCAGGCAATACAACTGCAGTGCAAGCTTCGTGTGACTTATAAG-CAAC------------GGCA---AATA------ |
| droVir3 | scaffold_13049:22941104-22941227 - | CCA-----AAATC----CGACA-----------TCT-CTGGAGCTGTTACGGAGGTGCATTGTAGTCGCATTGTCTGTC-------------------TCAAAT-TTGC-CCGCGGCAATACAACTGCAGTGCAAGCTCCGTGCAGTTCACACG-CAAC------------GTCG---GAAG------ |
| droMoj3 | scaffold_6680:17552824-17552948 - | CAG-----CATTCCGACA---------------TCT-TTGGAGCTGTGACGGAGGTGCATTGTAGTCGCATTGTCTGTA-------------------TCCTTTATTCT-CTGCGGCAATACAACTGCAGTGCAAGCTCCGTGCAGTTCACTCG-CAAC------------GTCG---GACA------ |
| droGri2 | scaffold_15110:18214439-18214590 - | CTCTGCACAGTTAGGGCAGACA-----------TCTCCAGGAGCTGTCACGAAGGTGCATTGTAGTCGCATTGTCTGTC-------------------CAAAAA-TTGC-ATGCGGCAATACAACTGCAGTGCAAGCTCCGTGCAGTTCACTAG-CAACAACTCTTTGATCGCTG---ATCGATCTGA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-19.5, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.2, p-value=0.009901 | dG=-18.9, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
| dG=-19.5, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.2, p-value=0.009901 | dG=-18.9, p-value=0.009901 |
|---|---|---|---|
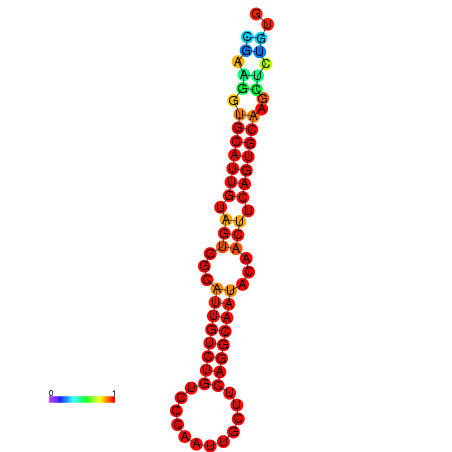 |
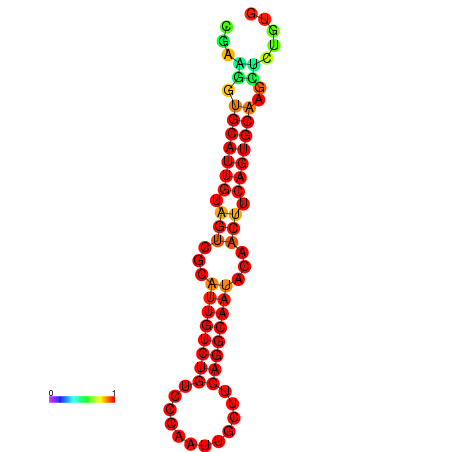 |
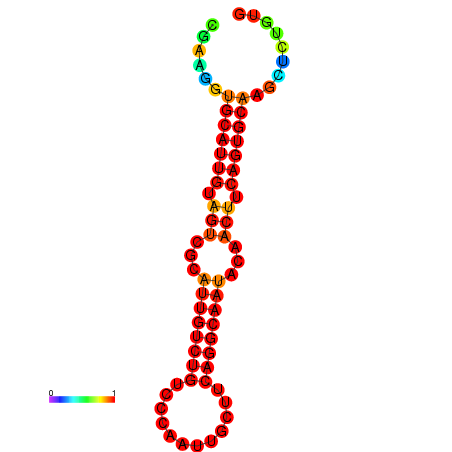 |
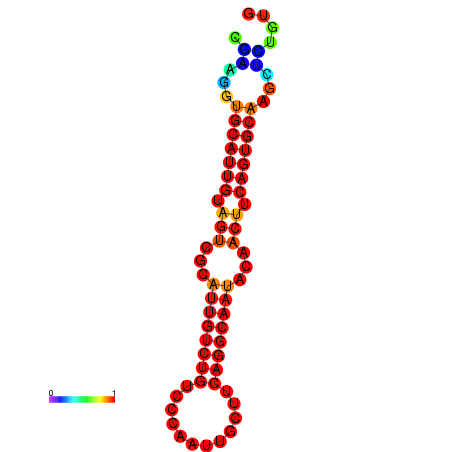 |
| dG=-19.5, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.2, p-value=0.009901 | dG=-18.9, p-value=0.009901 |
|---|---|---|---|
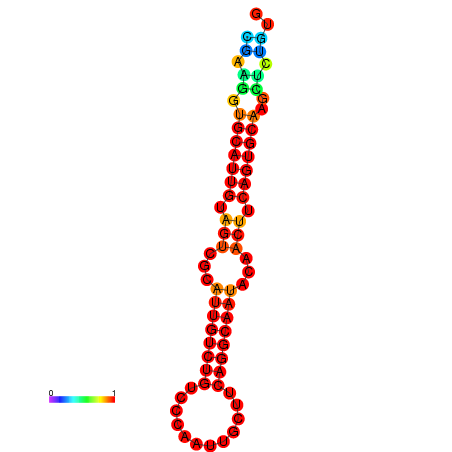 |
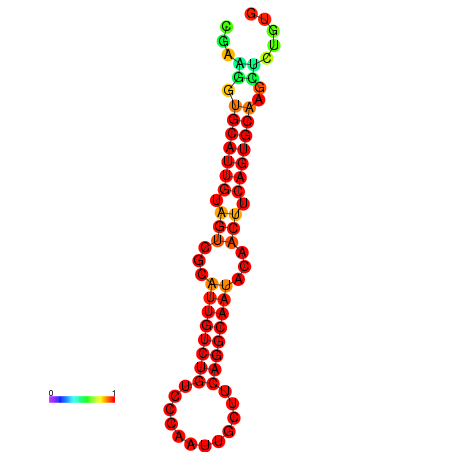 |
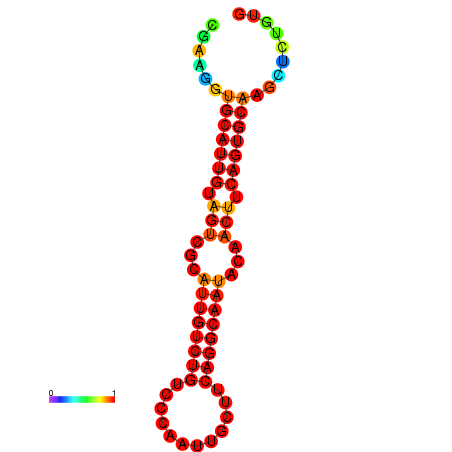 |
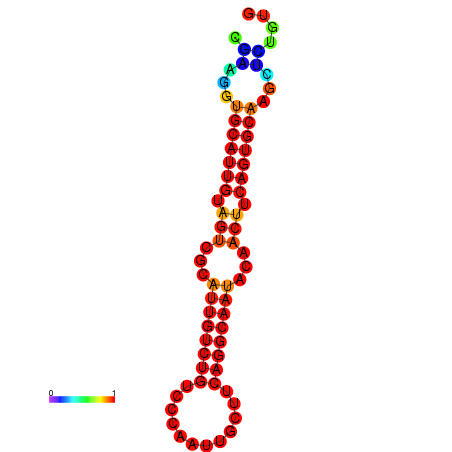 |
| dG=-19.5, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.2, p-value=0.009901 | dG=-18.9, p-value=0.009901 |
|---|---|---|---|
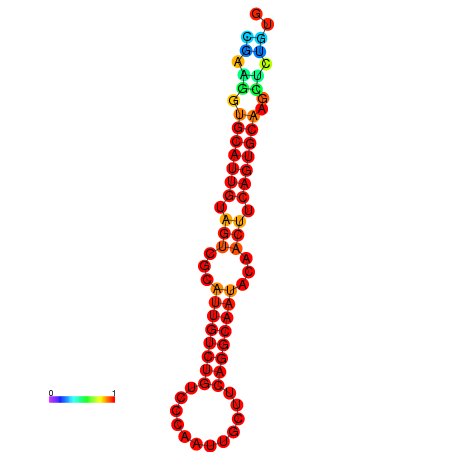 |
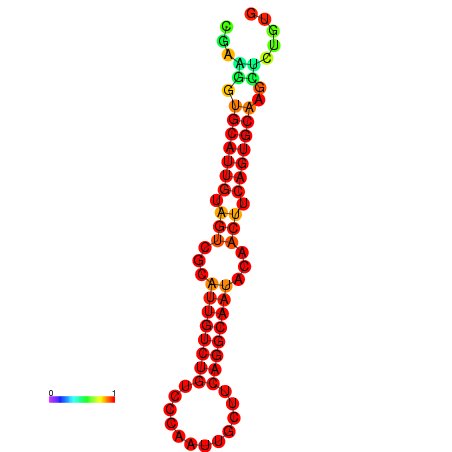 |
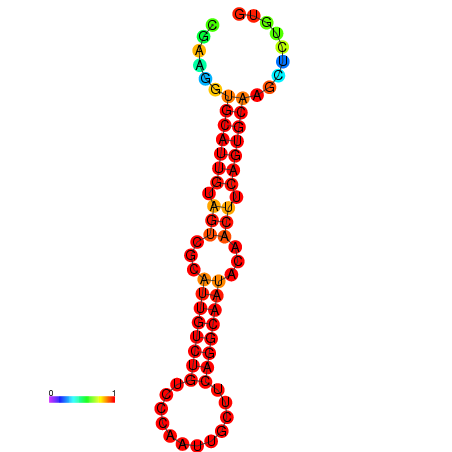 |
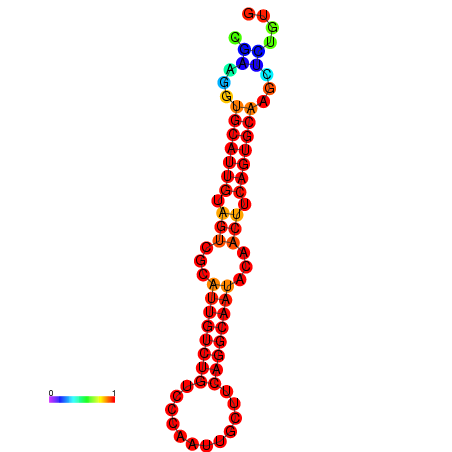 |
| dG=-19.5, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.2, p-value=0.009901 | dG=-18.9, p-value=0.009901 |
|---|---|---|---|
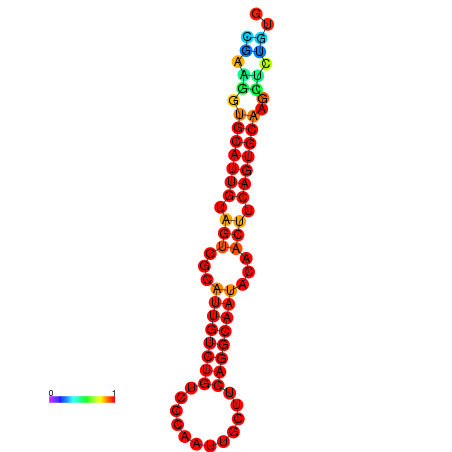 |
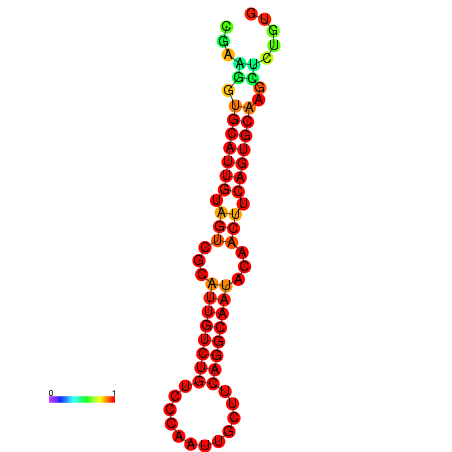 |
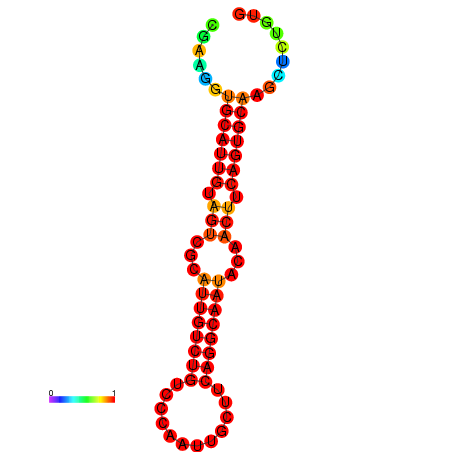 |
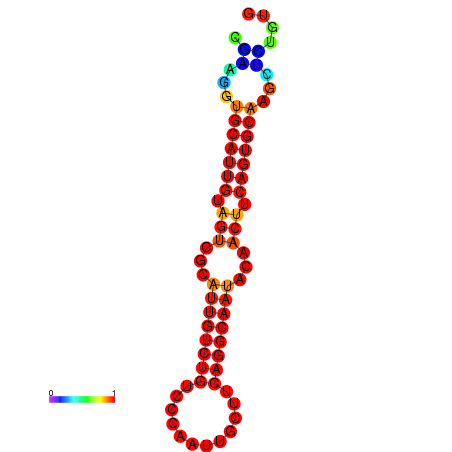 |
| dG=-28.1, p-value=0.009901 | dG=-27.4, p-value=0.009901 |
|---|---|
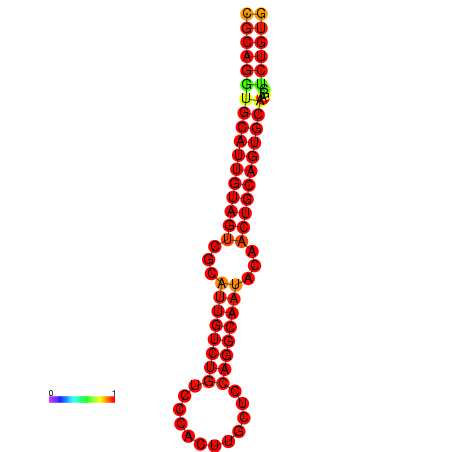 |
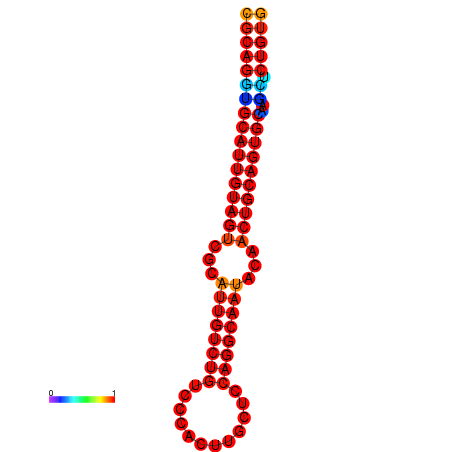 |
| dG=-28.6, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.9, p-value=0.009901 |
|---|---|---|
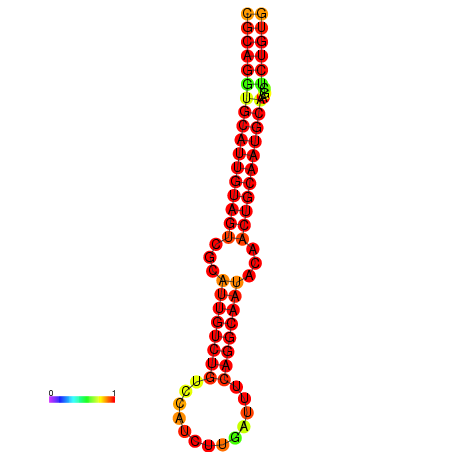 |
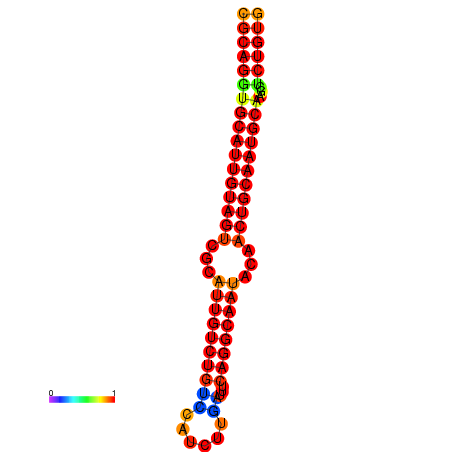 |
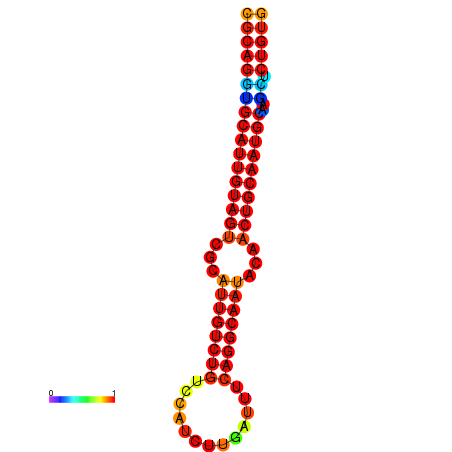 |
| dG=-28.6, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.9, p-value=0.009901 |
|---|---|---|
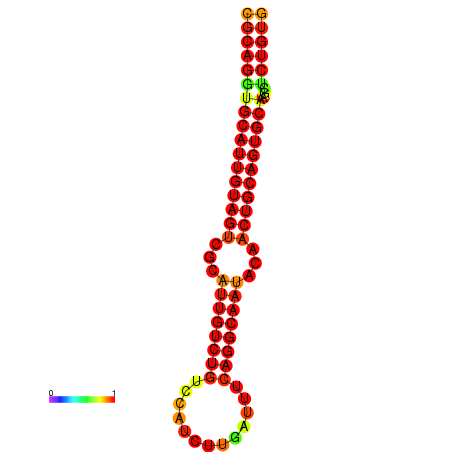 |
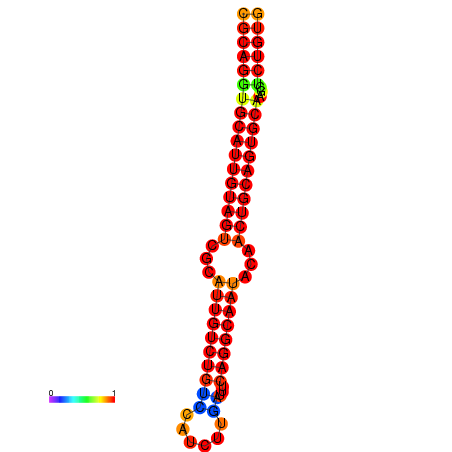 |
 |
| dG=-29.3, p-value=0.009901 | dG=-28.4, p-value=0.009901 |
|---|---|
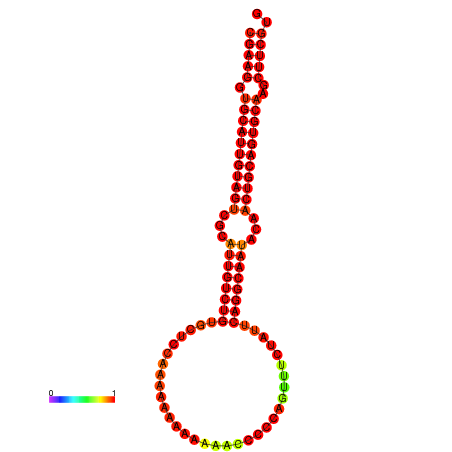 |
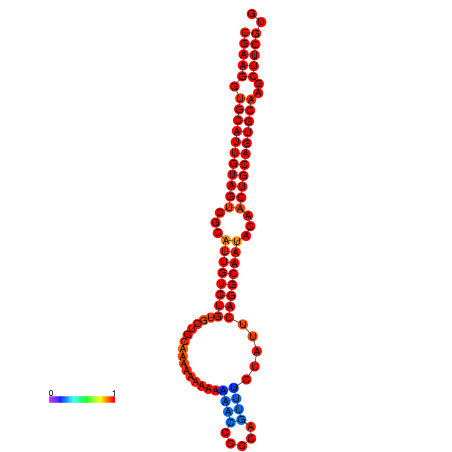 |
| dG=-29.0, p-value=0.009901 | dG=-28.6, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.2, p-value=0.009901 | dG=-28.1, p-value=0.009901 |
|---|---|---|---|---|
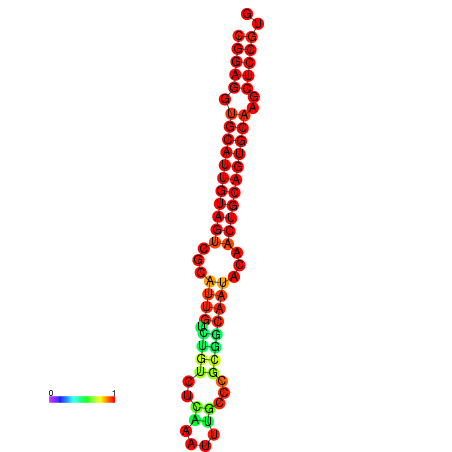 |
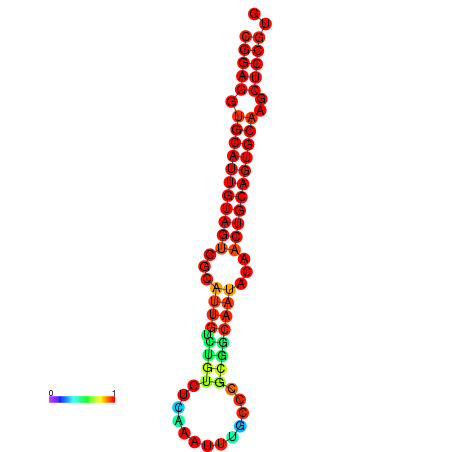 |
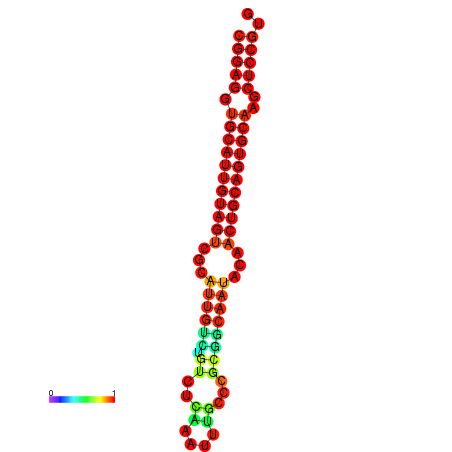 |
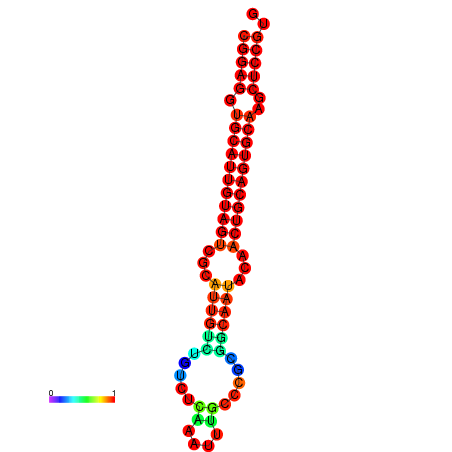 |
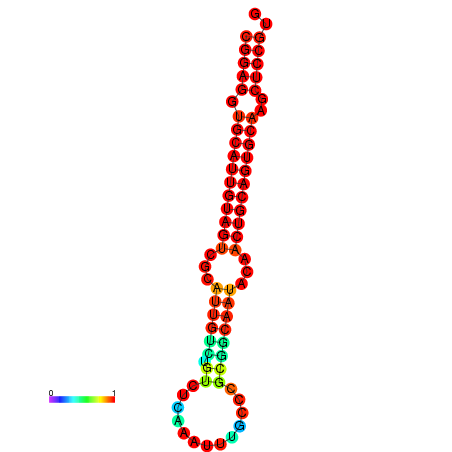 |
| dG=-29.9, p-value=0.009901 | dG=-29.4, p-value=0.009901 |
|---|---|
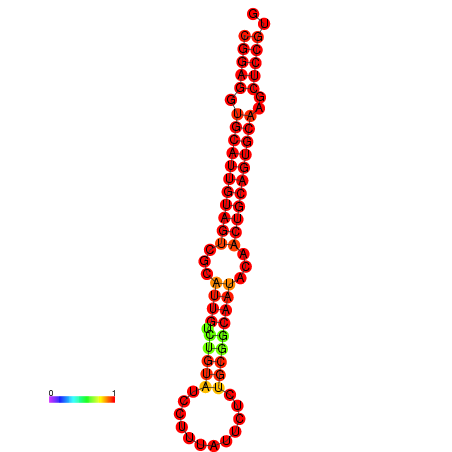 |
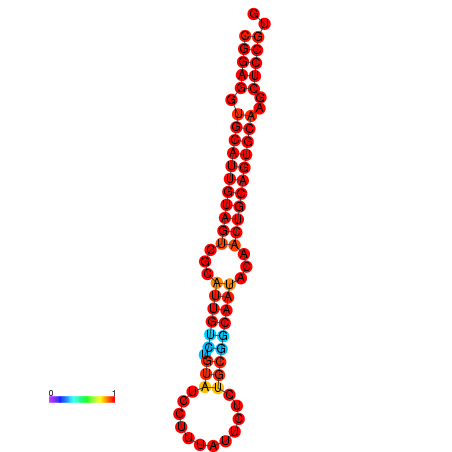 |
| dG=-22.5, p-value=0.009901 | dG=-22.3, p-value=0.009901 | dG=-22.1, p-value=0.009901 | dG=-22.0, p-value=0.009901 | dG=-21.9, p-value=0.009901 |
|---|---|---|---|---|
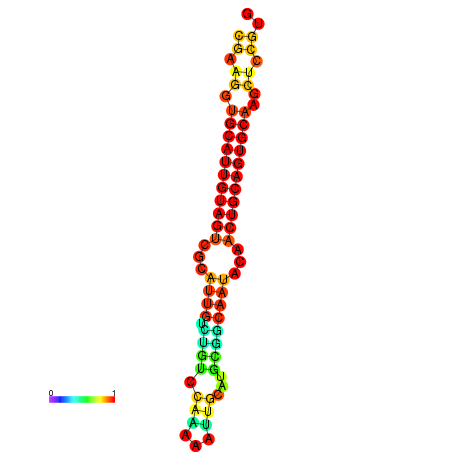 |
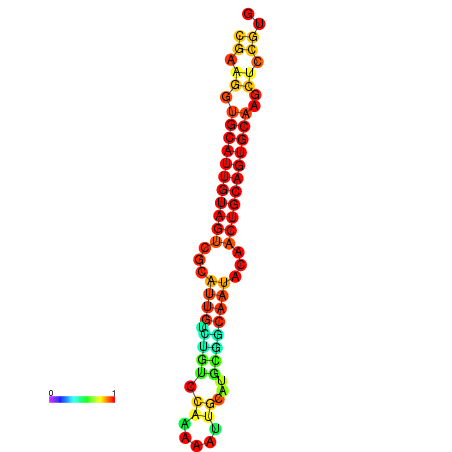 |
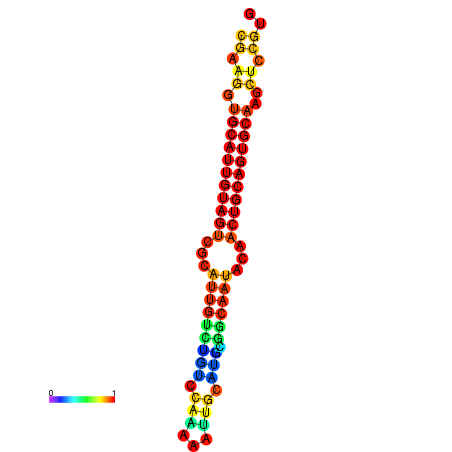 |
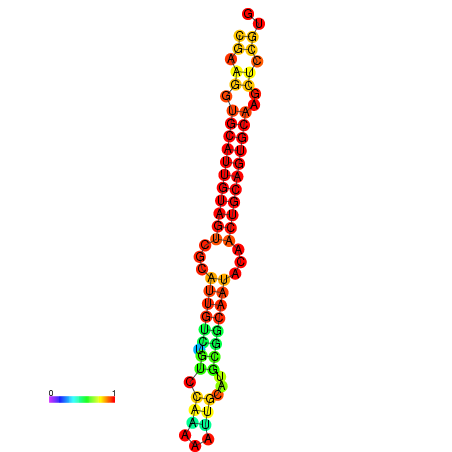 |
 |
Generated: 03/07/2013 at 05:22 PM