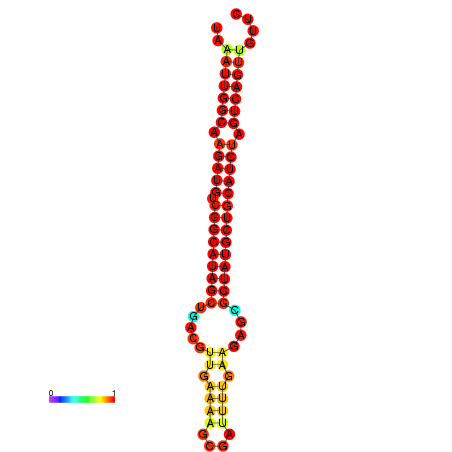
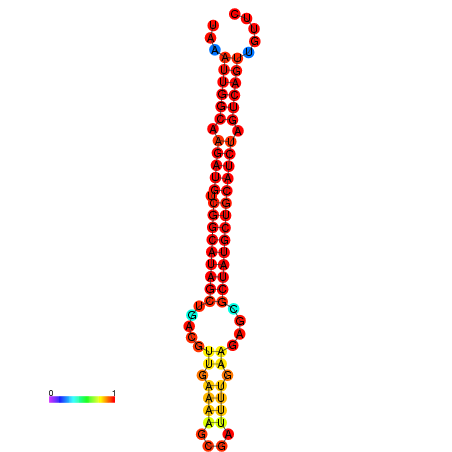
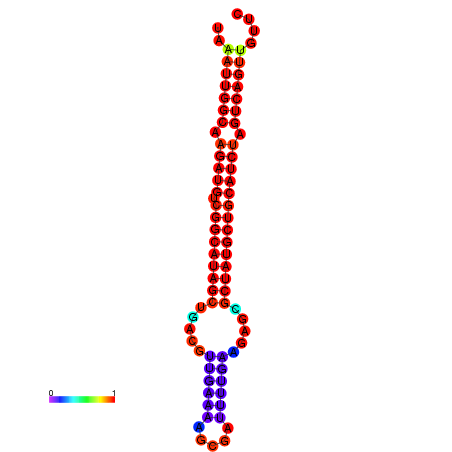
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:13671285-13671406 - | CGATC---------------GTAT--ATC---AACTCCGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGAAAAGCGATTTT--GAAGAGCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACAGTCAGCGAG---ATC------------------------------------------ |
| droSim1 | chr2R:12409842-12409963 - | CGATC---------------GTAT--AGC---ACCTCCGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGAAAAGCGATTTT--GAAGAGCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACAGTCAGCGAG---ATC------------------------------------------ |
| droSec1 | super_1:11168758-11168879 - | CGATC---------------GTAT--AGC---ACCTCCGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGAAAAGCGATTTT--GAAGAGCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACAGTCAGCGAG---ATC------------------------------------------ |
| droYak2 | chr2R:15512007-15512128 + | CGATC---------------GTAT--AGC---ACCTCTGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGAAAAGCGATTTT--GAAGATCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACACTCAGCGAG---ATC------------------------------------------ |
| droEre2 | scaffold_4845:7899182-7899303 - | CGATC---------------GTAT--AGC---ACCTCCGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGAAAAGCGCTTTT--AAAGAGCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACAGTCAGCGAG---ATC------------------------------------------ |
| droAna3 | scaffold_13266:10515593-10515725 + | CGATC---------------ATAT--AGCCAAGAATCTGTTGTT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGGAAAACGAATTT--TGAGAGCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACAGTCAACAAG---ATCGAAACTCT---------------------------------- |
| dp4 | chr3:16483204-16483318 - | CG---------------------T--ACC---GCATCTGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGAAGTTGAAAAGCGATCTT--TGAGAACGCTATGCTGCATCTAGTCAGT-TATTCAATGGAACAGTCAACA-G---ATC------------------------------------------ |
| droPer1 | super_4:278992-279106 + | CG---------------------T--ACC---GCATCTGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGAAGTTGAAAAGCGATCTT--TGAGAACGCTATGCTGCATCTAGTCAGT-TATTCAATGGAACAGTCAACA-G---ATC------------------------------------------ |
| droWil1 | scaffold_180700:4557862-4557981 - | CA-TC---------------ATCATTAT-----CATCTGTTTTTCGAATTGGCAAGATGTCGGCATAGCTGATA-------AT-ATTTTTAGAAAAGCGCTATGCTGCATCTAGTCAGTTTATTCAATAGAGCAATCGACAATATTATC------------------------------------------ |
| droVir3 | scaffold_12875:16937652-16937767 - | ---TC---------------GCCT--C-----GCATCTGTTGTT-CAATTGGCAAGATGTCGGCATAGCTGACGTTTGAAAGCGCATTT--TTATAGCGCTATGCTGCATCTAGTCAGT-TATTCAATGGAACAATCAACAAA---A-------------------------------------------- |
| droMoj3 | scaffold_6496:11065442-11065572 + | CTATATTTATAAATATTTCGGTTT--ATC---GCATCTATTGTT-GAATTGGCAAGATGTCGGCATAGCTGACGTTTCAAAGCGCAT-T--GCATAGCGCTATGCTGCATCTAGTCAGT-TATTCAATGGAACAGTCAACA-------------------------------------------------- |
| droGri2 | scaffold_15245:16476618-16476778 - | CTCTC---------------GCGA--TTC---TCATCTGTTGGT-GAATTGGCAAGATGTCGGCATAGCTGACGTTTGAAAGCGCATCT---TATAGCGCTATGCTGCATCTAGTCAGT-TATTCAATGGAACACTCAACA-T---ATCCATAC-CTGAAATGCTTAGAAACTTGGAGTATCTACGGATTA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:13671285-13671406 - | CGATC---------------GTAT--ATC---AACTCCGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGAAAAGCGATTTT--GAAGAGCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACAGTCAGCGAG---ATC------------------------------------------ |
| droSim1 | chr2R:12409842-12409963 - | CGATC---------------GTAT--AGC---ACCTCCGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGAAAAGCGATTTT--GAAGAGCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACAGTCAGCGAG---ATC------------------------------------------ |
| droSec1 | super_1:11168758-11168879 - | CGATC---------------GTAT--AGC---ACCTCCGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGAAAAGCGATTTT--GAAGAGCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACAGTCAGCGAG---ATC------------------------------------------ |
| droYak2 | chr2R:15512007-15512128 + | CGATC---------------GTAT--AGC---ACCTCTGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGAAAAGCGATTTT--GAAGATCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACACTCAGCGAG---ATC------------------------------------------ |
| droEre2 | scaffold_4845:7899182-7899303 - | CGATC---------------GTAT--AGC---ACCTCCGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGAAAAGCGCTTTT--AAAGAGCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACAGTCAGCGAG---ATC------------------------------------------ |
| droAna3 | scaffold_13266:10515593-10515725 + | CGATC---------------ATAT--AGCCAAGAATCTGTTGTT-AAATTGGCAAGATGTCGGCATAGCTGACGTTGGAAAACGAATTT--TGAGAGCGCTATGCTGCATCTAGTCAGT-TGTTCAATGGAACAGTCAACAAG---ATCGAAACTCT---------------------------------- |
| dp4 | chr3:16483204-16483318 - | CG---------------------T--ACC---GCATCTGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGAAGTTGAAAAGCGATCTT--TGAGAACGCTATGCTGCATCTAGTCAGT-TATTCAATGGAACAGTCAACA-G---ATC------------------------------------------ |
| droPer1 | super_4:278992-279106 + | CG---------------------T--ACC---GCATCTGTTGGT-AAATTGGCAAGATGTCGGCATAGCTGAAGTTGAAAAGCGATCTT--TGAGAACGCTATGCTGCATCTAGTCAGT-TATTCAATGGAACAGTCAACA-G---ATC------------------------------------------ |
| droWil1 | scaffold_180700:4557862-4557981 - | CA-TC---------------ATCATTAT-----CATCTGTTTTTCGAATTGGCAAGATGTCGGCATAGCTGATA-------AT-ATTTTTAGAAAAGCGCTATGCTGCATCTAGTCAGTTTATTCAATAGAGCAATCGACAATATTATC------------------------------------------ |
| droVir3 | scaffold_12875:16937652-16937767 - | ---TC---------------GCCT--C-----GCATCTGTTGTT-CAATTGGCAAGATGTCGGCATAGCTGACGTTTGAAAGCGCATTT--TTATAGCGCTATGCTGCATCTAGTCAGT-TATTCAATGGAACAATCAACAAA---A-------------------------------------------- |
| droMoj3 | scaffold_6496:11065442-11065572 + | CTATATTTATAAATATTTCGGTTT--ATC---GCATCTATTGTT-GAATTGGCAAGATGTCGGCATAGCTGACGTTTCAAAGCGCAT-T--GCATAGCGCTATGCTGCATCTAGTCAGT-TATTCAATGGAACAGTCAACA-------------------------------------------------- |
| droGri2 | scaffold_15245:16476618-16476778 - | CTCTC---------------GCGA--TTC---TCATCTGTTGGT-GAATTGGCAAGATGTCGGCATAGCTGACGTTTGAAAGCGCATCT---TATAGCGCTATGCTGCATCTAGTCAGT-TATTCAATGGAACACTCAACA-T---ATCCATAC-CTGAAATGCTTAGAAACTTGGAGTATCTACGGATTA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-29.5, p-value=0.009901 | dG=-28.8, p-value=0.009901 | dG=-28.6, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-29.5, p-value=0.009901 | dG=-28.8, p-value=0.009901 | dG=-28.6, p-value=0.009901 |
|---|---|---|
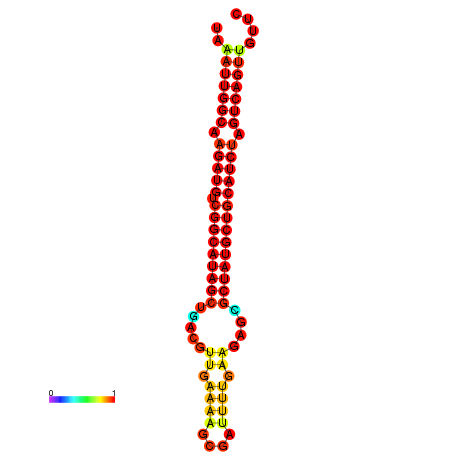 |
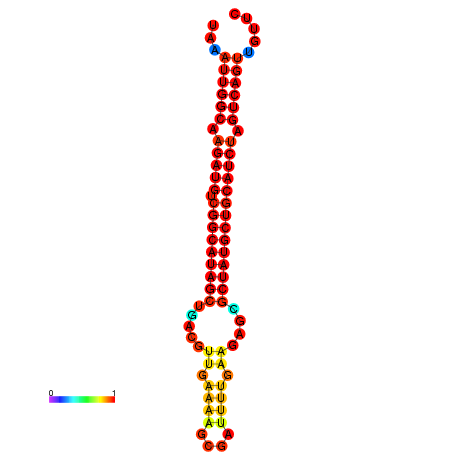 |
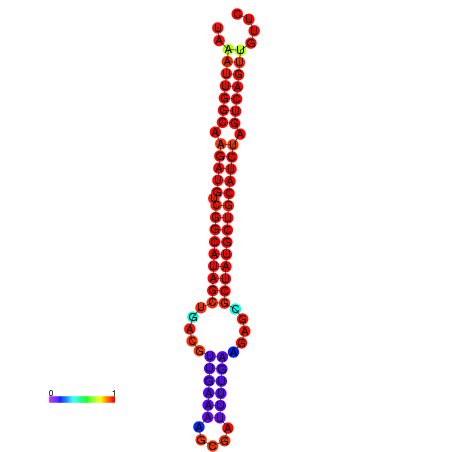 |
| dG=-29.5, p-value=0.009901 | dG=-28.8, p-value=0.009901 | dG=-28.6, p-value=0.009901 |
|---|---|---|
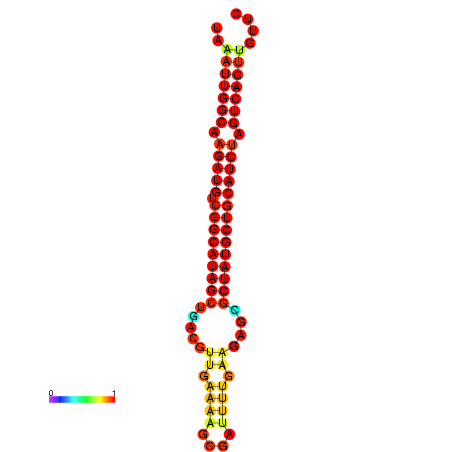 |
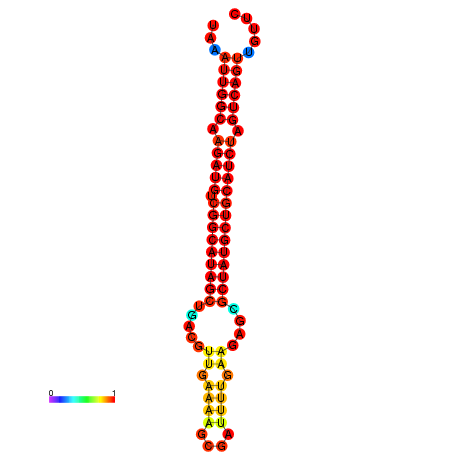 |
 |
| dG=-32.4, p-value=0.009901 | dG=-31.7, p-value=0.009901 |
|---|---|
 |
 |
| dG=-29.7, p-value=0.009901 | dG=-29.0, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.7, p-value=0.009901 | dG=-28.7, p-value=0.009901 |
|---|---|---|---|---|
 |
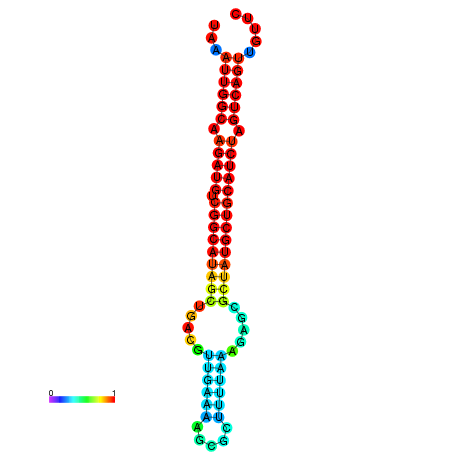 |
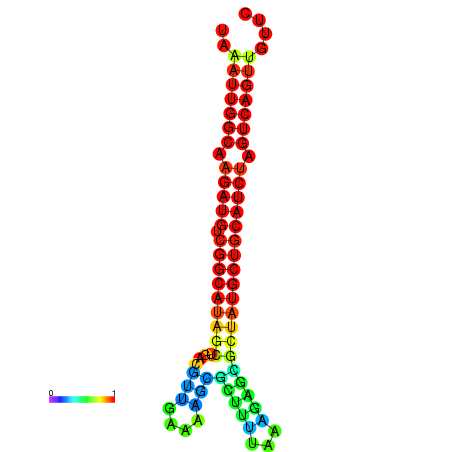 |
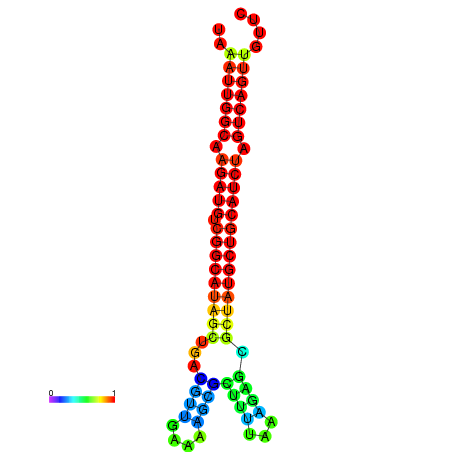 |
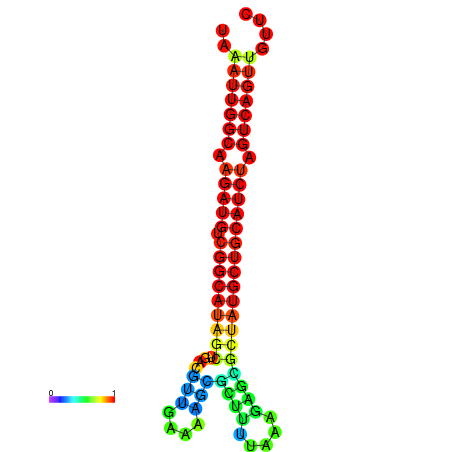 |
| dG=-28.2, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.5, p-value=0.009901 | dG=-27.4, p-value=0.009901 |
|---|---|---|---|---|
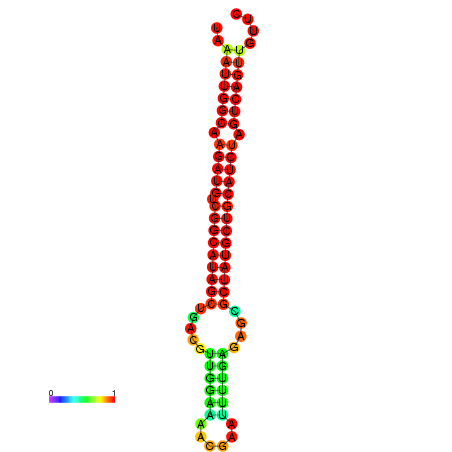 |
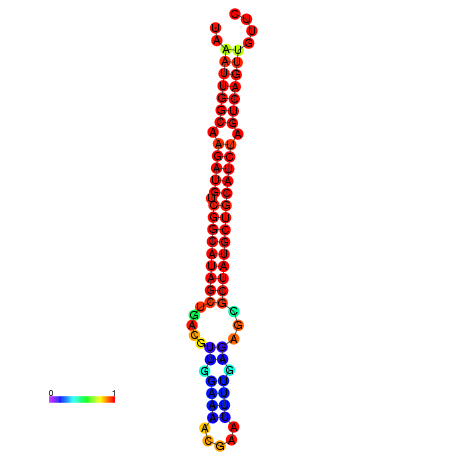 |
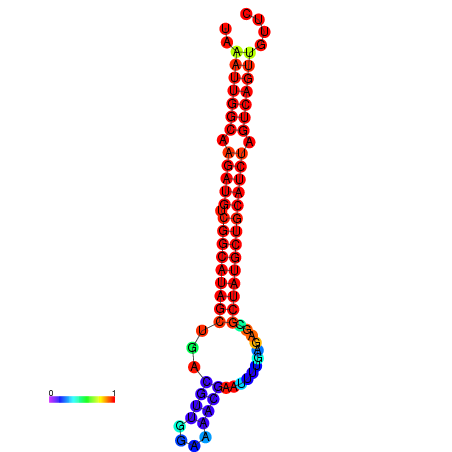 |
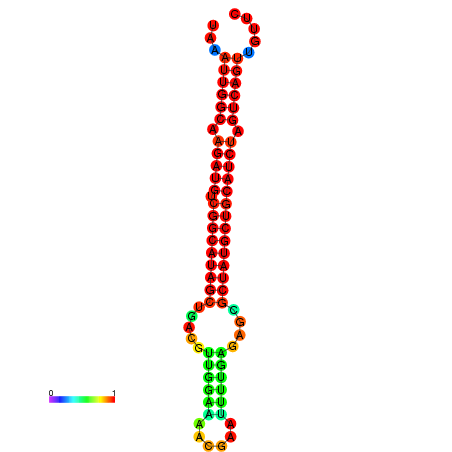 |
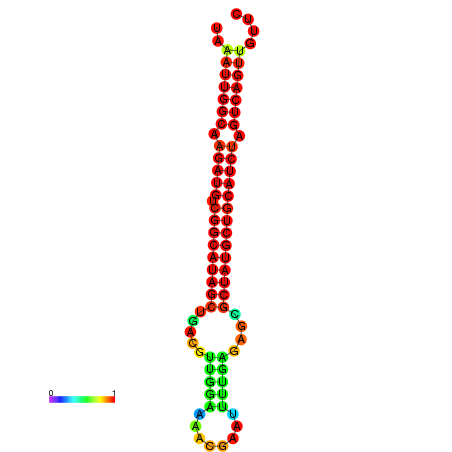 |
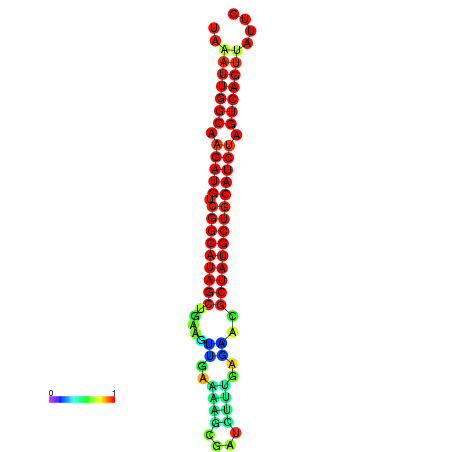
| dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.2, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
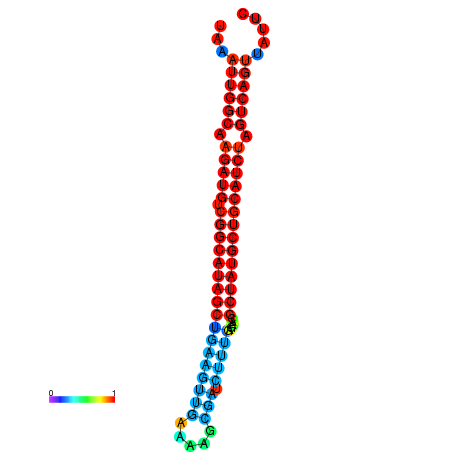 |
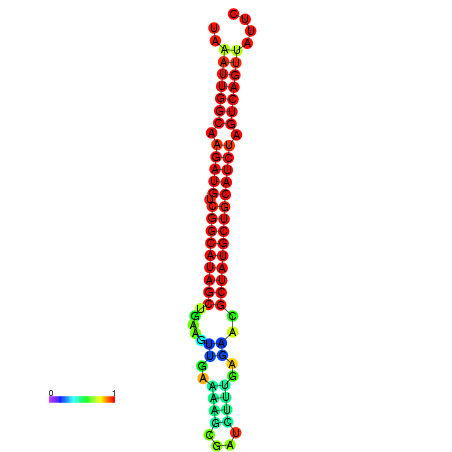
| dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.2, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-27.5, p-value=0.009901 | dG=-27.2, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.5, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
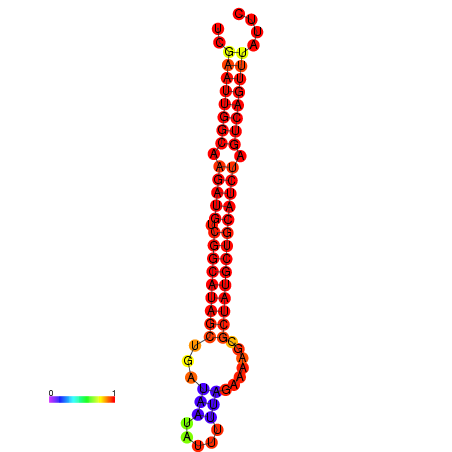 |
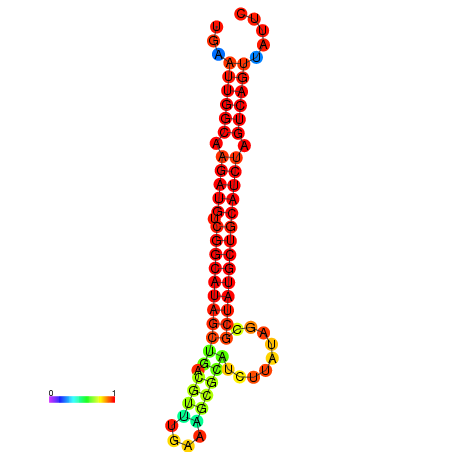
| dG=-28.5, p-value=0.009901 | dG=-28.1, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.9, p-value=0.009901 |
|---|---|---|---|---|
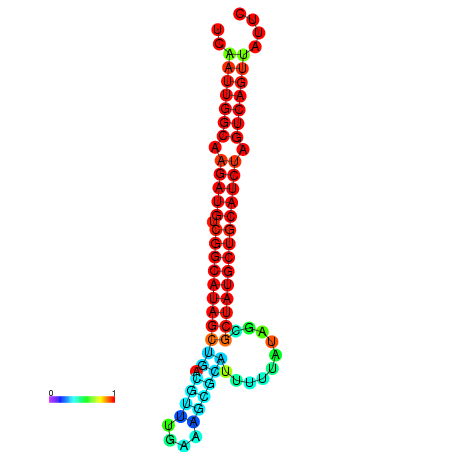 |
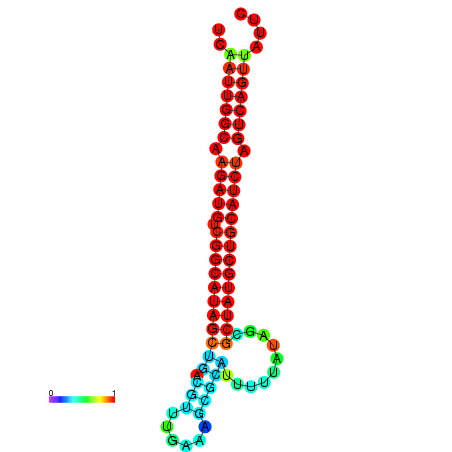 |
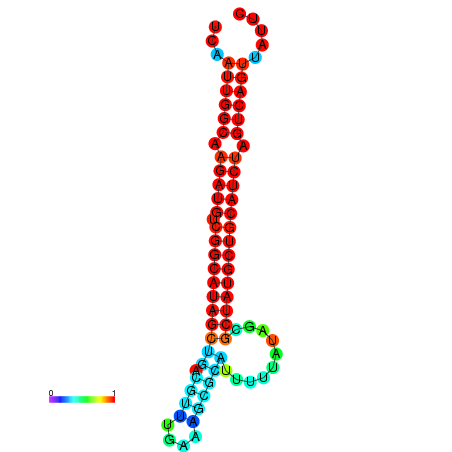 |
 |
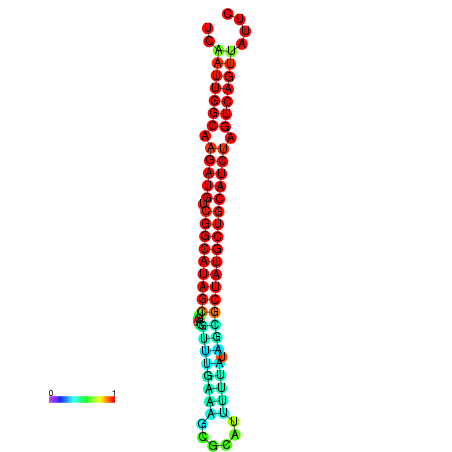 |
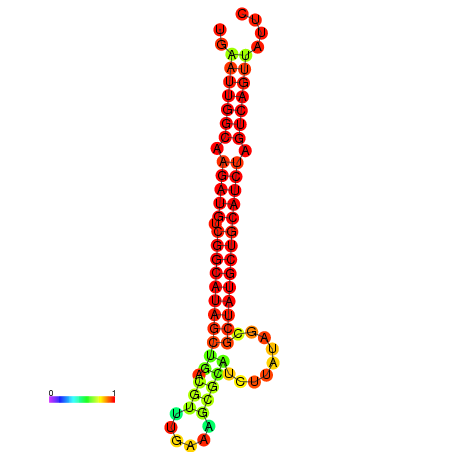
| dG=-28.8, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.1, p-value=0.009901 |
|---|---|---|
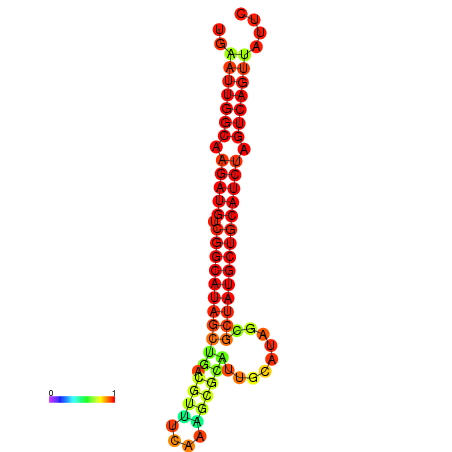 |
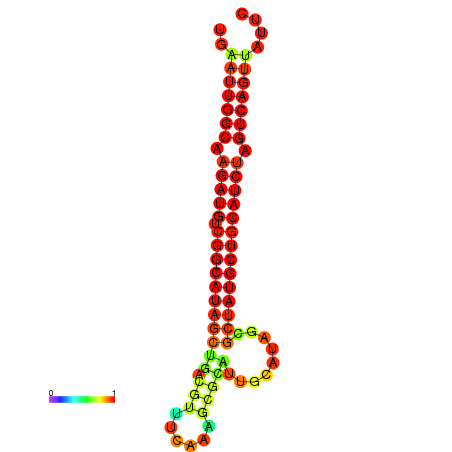 |
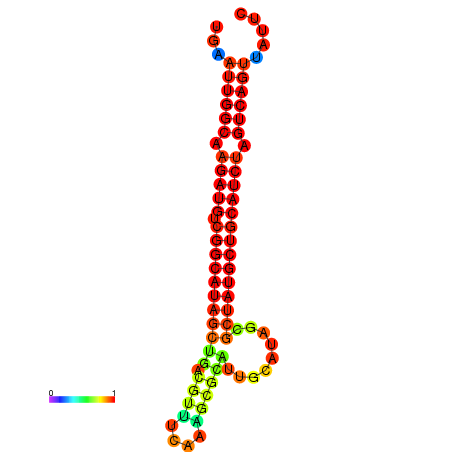 |
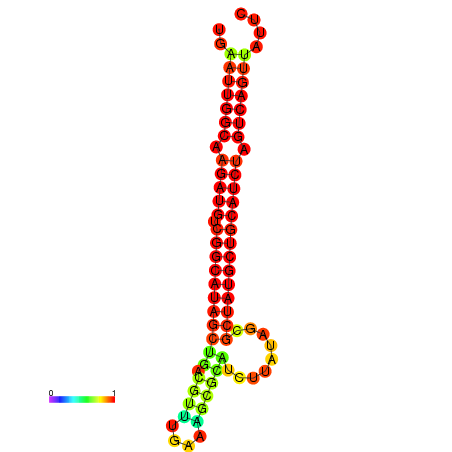
| dG=-28.8, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.1, p-value=0.009901 |
|---|---|---|
 |
 |
 |
Generated: 03/07/2013 at 05:21 PM