| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:6234009-6234106 + | G-------TCCTT--CGGTTTCAGTTTATGGGATACACACAGTTCAGTTTTGTCACAC-T--TCAAGCATCACTGGGCTTTGTTTATCTCATGAGAGCGAAATTTATTCA |
| droSim1 | chr3R:15159142-15159239 - | G-------TCCTT--CGGTTTCAGTTTATGGGATACACACAGTTCAGTTTTGTCACAC-T--TCAAGCATCACTGGGCTTTGTTTATCTCATGAGAGCGAAATTTATTCA |
| droSec1 | super_0:15709308-15709405 - | G-------TCCTT--CGGTTTCAGTTTATGGGATACACACAGTTCAGTTTTGTCACAC-T--TCAAGCATCACTGGGCTTTGTTTATCTCATGAGAGCGAAATTTATTCA |
| droYak2 | chr3R:10262524-10262621 + | G-------TCCTC--CGGTTTCAGTTTATGGGATACACACAGTTCAGTTTTGTCGCAC-T--ACAAGCATCACTGGGCTTTGTTTATCTCATGAGAGCGAAATTTGTTCA |
| droEre2 | scaffold_4770:15383893-15383990 - | G-------TCCTC--CGGTTTCAGTTTATGGGATACACACAGTTCAGTTTTGTCACAC-T--TCAAGCATCACTGGGCTTTGTTTATCTCATGAGAGCGAAATTTGTTCA |
| droAna3 | scaffold_13340:867198-867296 - | G-------TCCTC--CGGTTTCAATTTGTGGGATACACACGGTTCAGTTGTGTCACTTTT--GCGAGCATCACTGGGCTTTGTTTATCTCATAAGACTGAAATTTGTTCA |
| dp4 | chr2:7233334-7233438 - | CGTGTGCCTCTTT--TGGTTTCAATTTATGGGATGCACCAAGTTCAGTTTTGTCACAT-T--TCGAGCATCACTGGGCTTTGTTTATCTCATAAGAGCGAAATTGATTCA |
| droPer1 | super_0:1094016-1094113 - | C-------TCTTT--TGGTTTCAATTTATGGGATGCACCAAGTTCAGTTTTGTCACAT-T--TCGAGCATCACTGGGCTTTGTTTATCTCATAAGAGCGAAATTGATTCA |
| droWil1 | scaffold_181108:652-751 + | G-------TCAGA--CGGTTTCAATTTATGGGATACACCAAGTTCAGTTTTGTCGCAT-TCCACGAGCATCACTGGGCTTTGTTTATCTCATAAAGGAGAAATGAATACA |
| droVir3 | scaffold_13047:1194317-1194415 + | T-------GATTT--GGATTTCAATTTATGGGATACACCAAGTTCAGTTTTGTCAGATGT--CCAAGCATCACTGGGCTTTGTTTATCTCATAAGAGTGAAATTTGTTCA |
| droMoj3 | scaffold_6540:5225327-5225427 + | G-------TGATTAAGGATTTCAATTTATGGGATACACTAAGTTCAGTTTTGTCAGATGT--CCAAGCATCACTGGGCTTTGTTTATCTCATAAGAGCGAAATTTGTTCA |
| droGri2 | scaffold_14906:5077291-5077389 - | G-------ATTTC--GGATTTCAATTTATGGGATACACCAAGTTCAGTTTTGTCAGATGT--CCAAGCATCACTGGGCTTTGTTTATCTCATAAGAGCGAAATTAGTTCA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:6234009-6234106 + | G-------TCCTT--CGGTTTCAGTTTATGGGATACACACAGTTCAGTTTTGTCACAC-T--TCAAGCATCACTGGGCTTTGTTTATCTCATGAGAGCGAAATTTATTCA |
| droSim1 | chr3R:15159142-15159239 - | G-------TCCTT--CGGTTTCAGTTTATGGGATACACACAGTTCAGTTTTGTCACAC-T--TCAAGCATCACTGGGCTTTGTTTATCTCATGAGAGCGAAATTTATTCA |
| droSec1 | super_0:15709308-15709405 - | G-------TCCTT--CGGTTTCAGTTTATGGGATACACACAGTTCAGTTTTGTCACAC-T--TCAAGCATCACTGGGCTTTGTTTATCTCATGAGAGCGAAATTTATTCA |
| droYak2 | chr3R:10262524-10262621 + | G-------TCCTC--CGGTTTCAGTTTATGGGATACACACAGTTCAGTTTTGTCGCAC-T--ACAAGCATCACTGGGCTTTGTTTATCTCATGAGAGCGAAATTTGTTCA |
| droEre2 | scaffold_4770:15383893-15383990 - | G-------TCCTC--CGGTTTCAGTTTATGGGATACACACAGTTCAGTTTTGTCACAC-T--TCAAGCATCACTGGGCTTTGTTTATCTCATGAGAGCGAAATTTGTTCA |
| droAna3 | scaffold_13340:867198-867296 - | G-------TCCTC--CGGTTTCAATTTGTGGGATACACACGGTTCAGTTGTGTCACTTTT--GCGAGCATCACTGGGCTTTGTTTATCTCATAAGACTGAAATTTGTTCA |
| dp4 | chr2:7233334-7233438 - | CGTGTGCCTCTTT--TGGTTTCAATTTATGGGATGCACCAAGTTCAGTTTTGTCACAT-T--TCGAGCATCACTGGGCTTTGTTTATCTCATAAGAGCGAAATTGATTCA |
| droPer1 | super_0:1094016-1094113 - | C-------TCTTT--TGGTTTCAATTTATGGGATGCACCAAGTTCAGTTTTGTCACAT-T--TCGAGCATCACTGGGCTTTGTTTATCTCATAAGAGCGAAATTGATTCA |
| droWil1 | scaffold_181108:652-751 + | G-------TCAGA--CGGTTTCAATTTATGGGATACACCAAGTTCAGTTTTGTCGCAT-TCCACGAGCATCACTGGGCTTTGTTTATCTCATAAAGGAGAAATGAATACA |
| droVir3 | scaffold_13047:1194317-1194415 + | T-------GATTT--GGATTTCAATTTATGGGATACACCAAGTTCAGTTTTGTCAGATGT--CCAAGCATCACTGGGCTTTGTTTATCTCATAAGAGTGAAATTTGTTCA |
| droMoj3 | scaffold_6540:5225327-5225427 + | G-------TGATTAAGGATTTCAATTTATGGGATACACTAAGTTCAGTTTTGTCAGATGT--CCAAGCATCACTGGGCTTTGTTTATCTCATAAGAGCGAAATTTGTTCA |
| droGri2 | scaffold_14906:5077291-5077389 - | G-------ATTTC--GGATTTCAATTTATGGGATACACCAAGTTCAGTTTTGTCAGATGT--CCAAGCATCACTGGGCTTTGTTTATCTCATAAGAGCGAAATTAGTTCA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-24.2, p-value=0.009901 |
|---|
 |
| dG=-24.2, p-value=0.009901 |
|---|
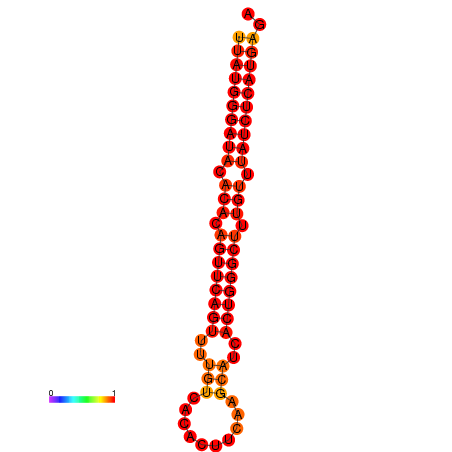
 |
| dG=-24.2, p-value=0.009901 |
|---|
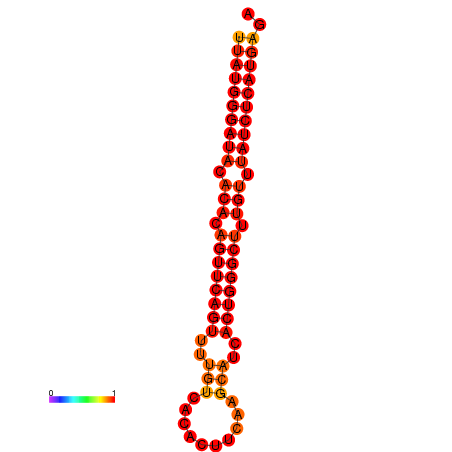
 |
| dG=-24.2, p-value=0.009901 | dG=-23.4, p-value=0.009901 |
|---|---|
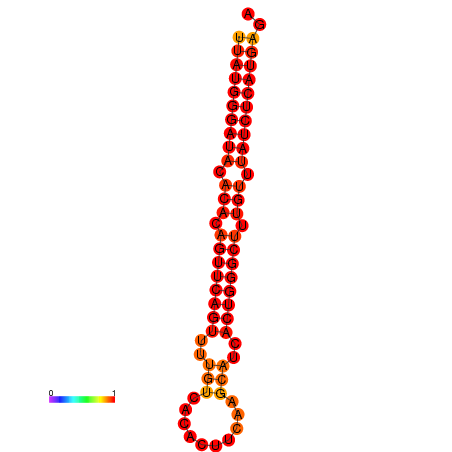
 |
 |
| dG=-24.2, p-value=0.009901 |
|---|
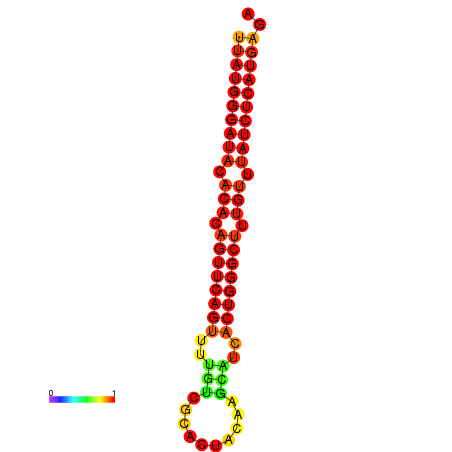
 |
| dG=-25.9, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-25.0, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-23.4, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.4, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
| dG=-23.4, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-22.5, p-value=0.009901 | dG=-22.4, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
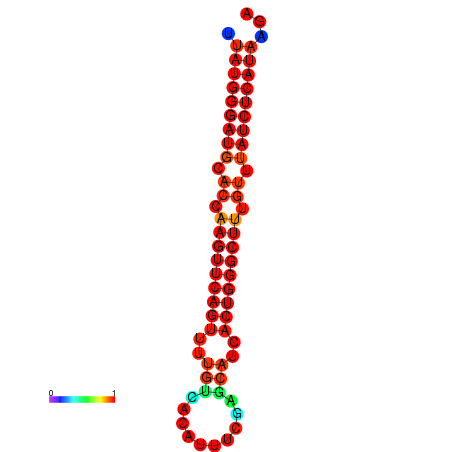 |
| dG=-25.7, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-24.8, p-value=0.009901 |
|---|---|---|
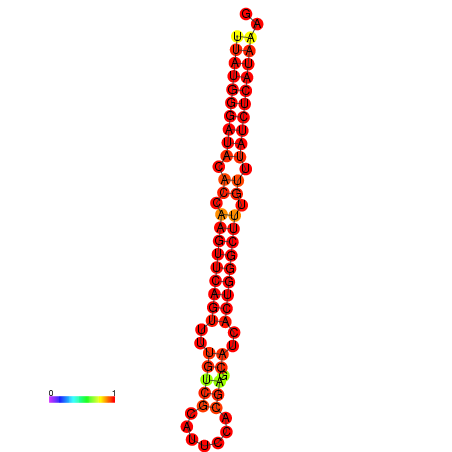 |
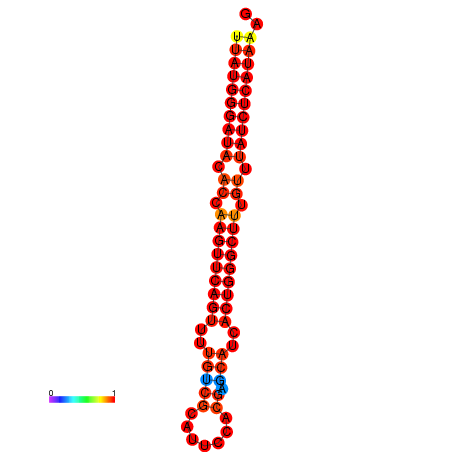 |
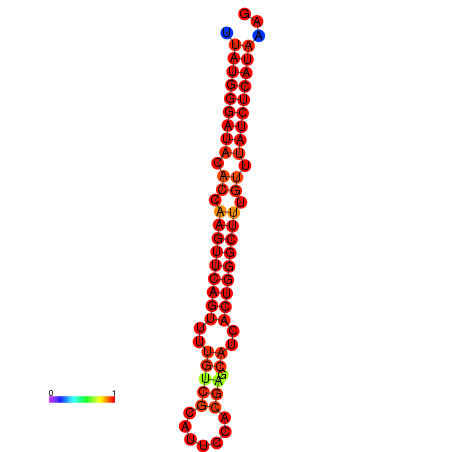 |
| dG=-24.6, p-value=0.009901 | dG=-23.7, p-value=0.009901 |
|---|---|
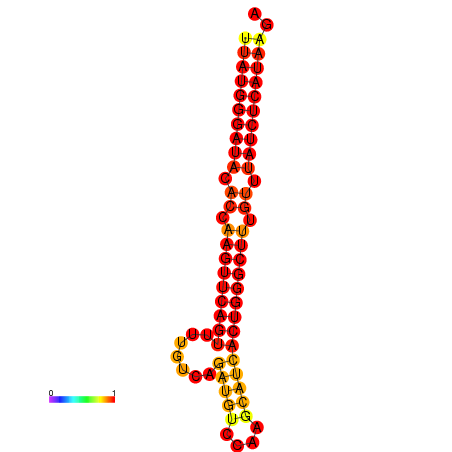 |
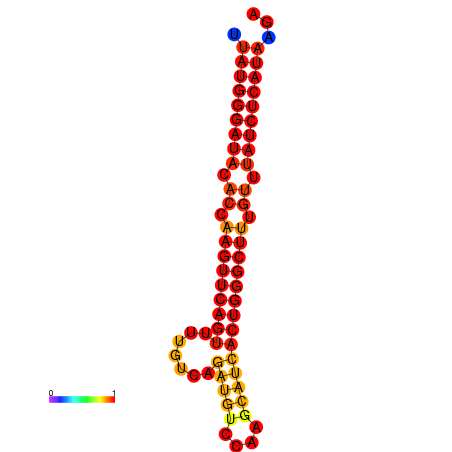 |
| dG=-24.6, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-23.7, p-value=0.009901 |
|---|---|---|
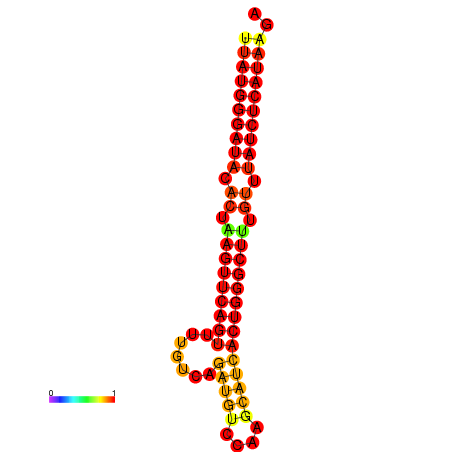 |
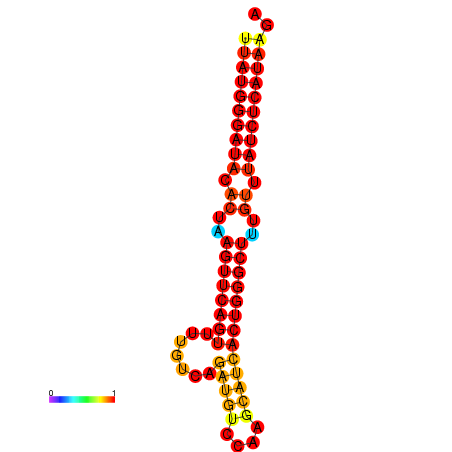 |
 |
| dG=-24.6, p-value=0.009901 | dG=-23.7, p-value=0.009901 |
|---|---|
 |
 |
Generated: 03/07/2013 at 05:20 PM