| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:5916833-5916954 + | AA----GC------GAATTTTCCA-------------TATGCAACTGCCATTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CATCCT-TGCTGCACCA-------------CA- |
| droSim1 | chr3R:15459127-15459249 - | AA----GC------GAATTTTCCA-------------TATGCAACTGCCATTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CATCCTTTGCTGCTCCA-------------CA- |
| droSec1 | super_0:15996573-15996694 - | AA----GC------GAATTTTCCA-------------TATGCAACTGCCATTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CATCCT-TGCTGCTCCA-------------CA- |
| droYak2 | chr3R:9963959-9964079 + | AA----GC------GAATTTTCCA-------------TATGCAACTGCCATTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CGTCCT-TGCTGCTCCAG--------------- |
| droEre2 | scaffold_4770:15683457-15683577 - | AA----GC------GAATTTTCCA-------------TATGCAACTGCCATTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CGTCCT-TGCTGCTCCAG--------------- |
| droAna3 | scaffold_13340:6479980-6480089 + | AA----GC------TCATTT---G-------------TGTGCATCGGCTGCTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TTG-CATCCT-TTC----------------------- |
| dp4 | chr2:5029980-5030116 - | AA----GC---------TCTTCCGTGTCTCGTGTCTCTGTGCAACTGCCGTTGGGATAC-ACCCTGTGCTCGCTTTGAATATGGTGCAAGCAAGTGAACACAGCTGGTGGTATCCAATGGCCGTT--CTG-CAACAA-TGCAACATCAGC---------AACAA |
| droPer1 | super_19:735499-735629 - | AA----GC---------TCTTCCGTGTCTCGTGTCTCTGTGCAACTGCCGTTGGGATAC-ACCCTGTGCTCGCTTTGAATATGGTGCAAGCAAGTGAACACAGCTGGTGGTATCCAATGGCCGTT--CTG-CAACAA-TGCAACATCAG--------------- |
| droWil1 | scaffold_181089:11180051-11180176 - | AA----GCTTTCTAGATTCTTC---------------TGTGCAACTGCCACTGGGATAC-ACCCTGTGCTCGCTTTGAATATTGTGCAAACAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTTTTTTG-CATCCA-TGCAACACCA---------------- |
| droVir3 | scaffold_12855:2698510-2698638 - | AA----GC---------TTTTTC--------------AGTGCAATTGCCGCTGGGATAC-ACCCTGTGCTCGCTTTGAATT--GTGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT---GGCAGCCAA-TGCAACAATTGCAACAACTGCAACA- |
| droMoj3 | scaffold_6540:11898843-11898966 - | GAGTAAGC----------TTTCT-----------CTCTGTGCAATTGCCGCTGGGATAC-ACCCTGTGCTCGCTTTGAGTT-TATGT-TGCAAGTGAACACAGCTGGTGGTATCCAGAGGCCGTT---GACAGCCAA-TGCAACAACAGC---------A--A- |
| droGri2 | scaffold_14624:2253109-2253242 + | AAA-AAGC---------TTTTCGC-------------TGTGCAATTGCCACTGGGATATTGCCCTGTGCTCGCTTTGAATATTGTGTAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CGCACA-TGCAACAACAACAACAAGAGCA--A- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:5916833-5916954 + | AA----GC------GAATTTTCCA-------------TATGCAACTGCCATTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CATCCT-TGCTGCACCA-------------CA- |
| droSim1 | chr3R:15459127-15459249 - | AA----GC------GAATTTTCCA-------------TATGCAACTGCCATTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CATCCTTTGCTGCTCCA-------------CA- |
| droSec1 | super_0:15996573-15996694 - | AA----GC------GAATTTTCCA-------------TATGCAACTGCCATTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CATCCT-TGCTGCTCCA-------------CA- |
| droYak2 | chr3R:9963959-9964079 + | AA----GC------GAATTTTCCA-------------TATGCAACTGCCATTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CGTCCT-TGCTGCTCCAG--------------- |
| droEre2 | scaffold_4770:15683457-15683577 - | AA----GC------GAATTTTCCA-------------TATGCAACTGCCATTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CGTCCT-TGCTGCTCCAG--------------- |
| droAna3 | scaffold_13340:6479980-6480089 + | AA----GC------TCATTT---G-------------TGTGCATCGGCTGCTGGGATAC-ACCCTGTGCTCGCTTTGAATGAAATGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TTG-CATCCT-TTC----------------------- |
| dp4 | chr2:5029980-5030116 - | AA----GC---------TCTTCCGTGTCTCGTGTCTCTGTGCAACTGCCGTTGGGATAC-ACCCTGTGCTCGCTTTGAATATGGTGCAAGCAAGTGAACACAGCTGGTGGTATCCAATGGCCGTT--CTG-CAACAA-TGCAACATCAGC---------AACAA |
| droPer1 | super_19:735499-735629 - | AA----GC---------TCTTCCGTGTCTCGTGTCTCTGTGCAACTGCCGTTGGGATAC-ACCCTGTGCTCGCTTTGAATATGGTGCAAGCAAGTGAACACAGCTGGTGGTATCCAATGGCCGTT--CTG-CAACAA-TGCAACATCAG--------------- |
| droWil1 | scaffold_181089:11180051-11180176 - | AA----GCTTTCTAGATTCTTC---------------TGTGCAACTGCCACTGGGATAC-ACCCTGTGCTCGCTTTGAATATTGTGCAAACAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTTTTTTG-CATCCA-TGCAACACCA---------------- |
| droVir3 | scaffold_12855:2698510-2698638 - | AA----GC---------TTTTTC--------------AGTGCAATTGCCGCTGGGATAC-ACCCTGTGCTCGCTTTGAATT--GTGCAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT---GGCAGCCAA-TGCAACAATTGCAACAACTGCAACA- |
| droMoj3 | scaffold_6540:11898843-11898966 - | GAGTAAGC----------TTTCT-----------CTCTGTGCAATTGCCGCTGGGATAC-ACCCTGTGCTCGCTTTGAGTT-TATGT-TGCAAGTGAACACAGCTGGTGGTATCCAGAGGCCGTT---GACAGCCAA-TGCAACAACAGC---------A--A- |
| droGri2 | scaffold_14624:2253109-2253242 + | AAA-AAGC---------TTTTCGC-------------TGTGCAATTGCCACTGGGATATTGCCCTGTGCTCGCTTTGAATATTGTGTAAGCAAGTGAACACAGCTGGTGGTATCCAGTGGCCGTT--TGG-CGCACA-TGCAACAACAACAACAAGAGCA--A- |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-29.3, p-value=0.009901 | dG=-29.3, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
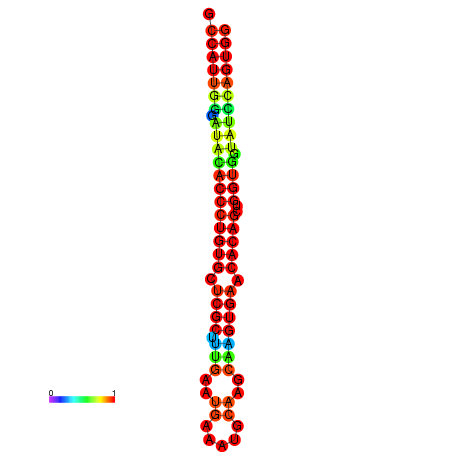
| dG=-29.3, p-value=0.009901 | dG=-29.3, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.9, p-value=0.009901 |
|---|---|---|---|---|
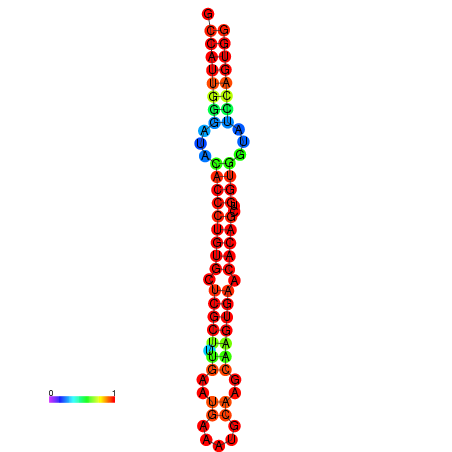 |
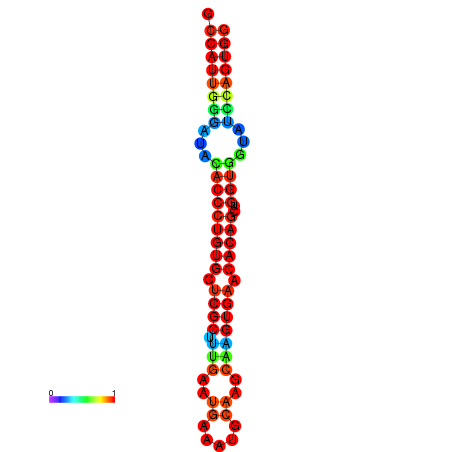 |
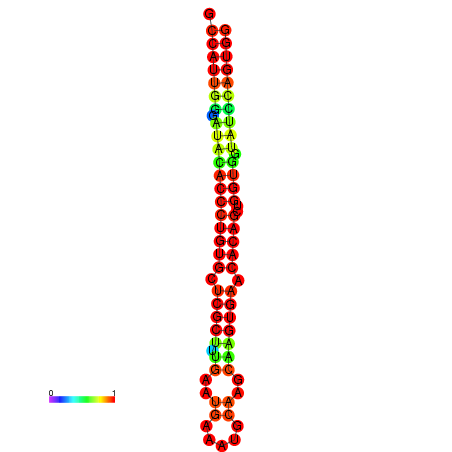 |
 |
 |
| dG=-29.3, p-value=0.009901 | dG=-29.3, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
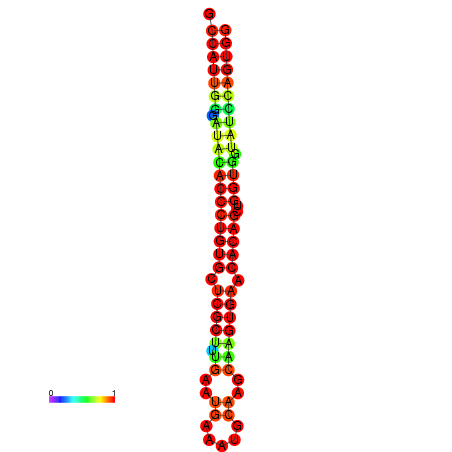 |
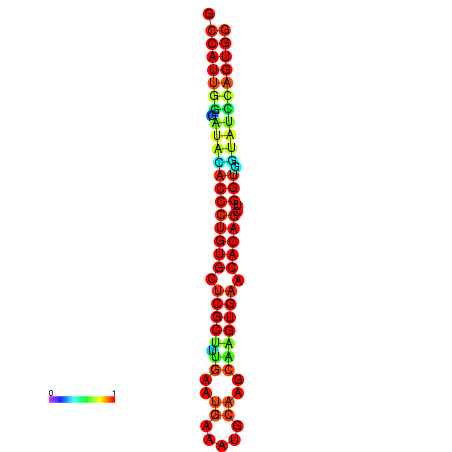 |
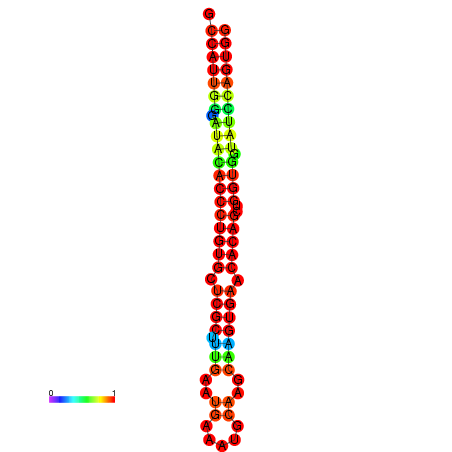 |
| dG=-29.3, p-value=0.009901 | dG=-29.3, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.9, p-value=0.009901 |
|---|---|---|---|---|
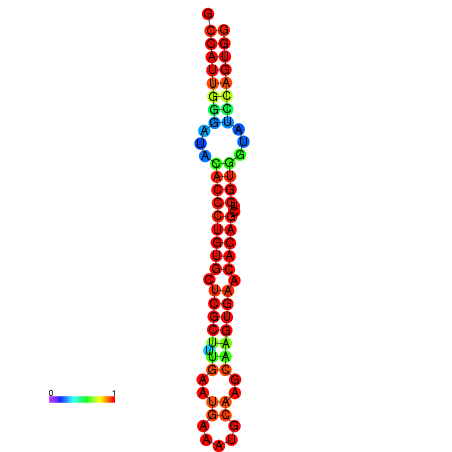 |
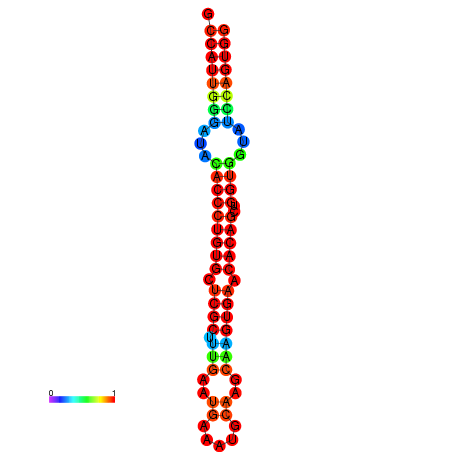 |
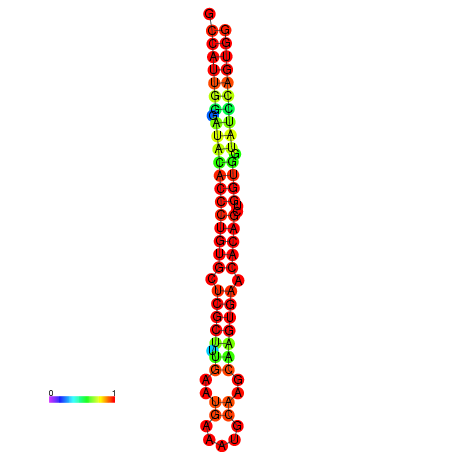 |
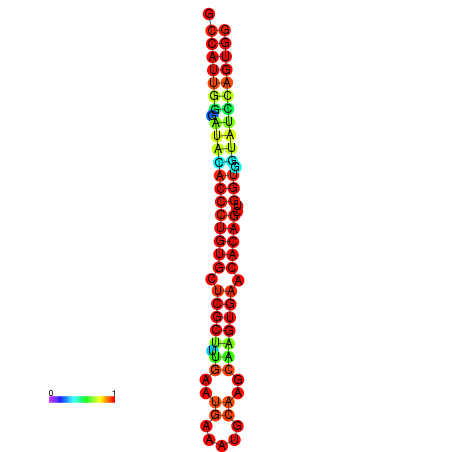 |
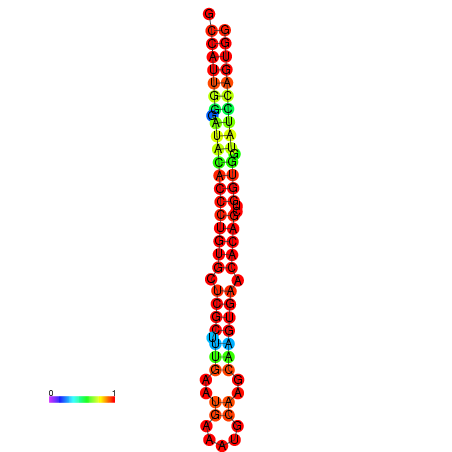 |
| dG=-29.3, p-value=0.009901 | dG=-29.3, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.9, p-value=0.009901 | dG=-28.9, p-value=0.009901 |
|---|---|---|---|---|
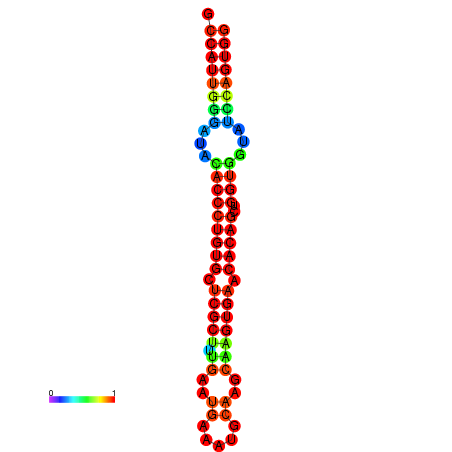 |
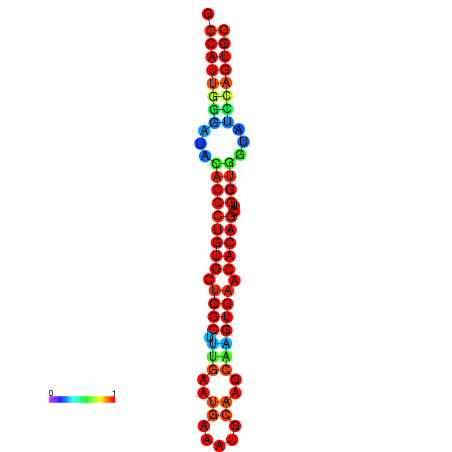 |
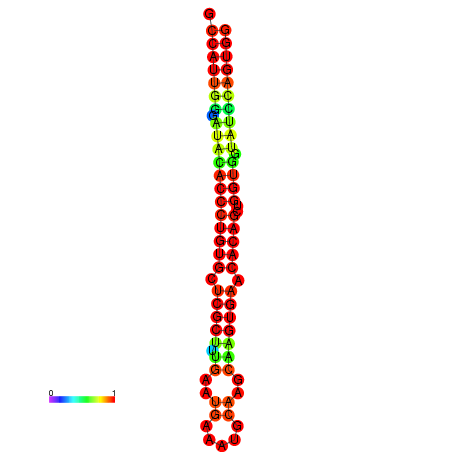 |
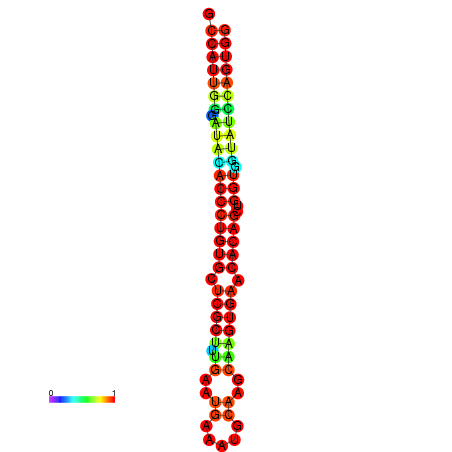 |
 |
| dG=-28.3, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-26.7, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
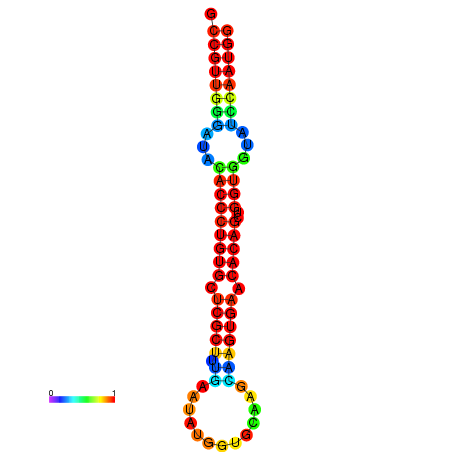 |
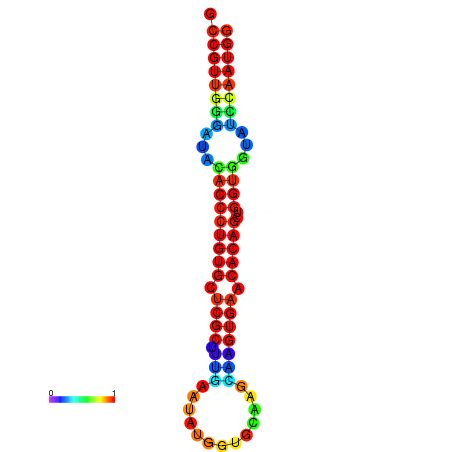 |
| dG=-26.7, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.4, p-value=0.009901 |
|---|---|---|---|---|
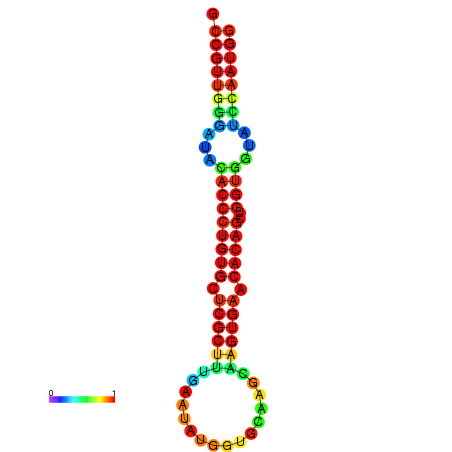 |
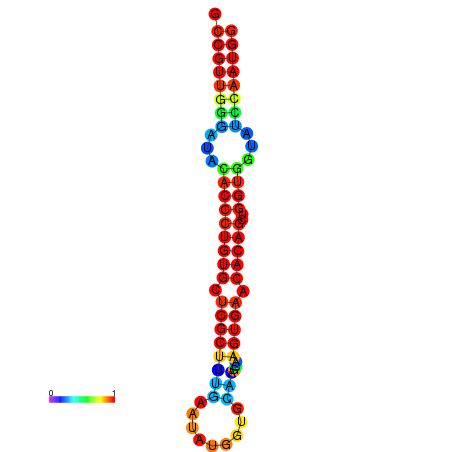 |
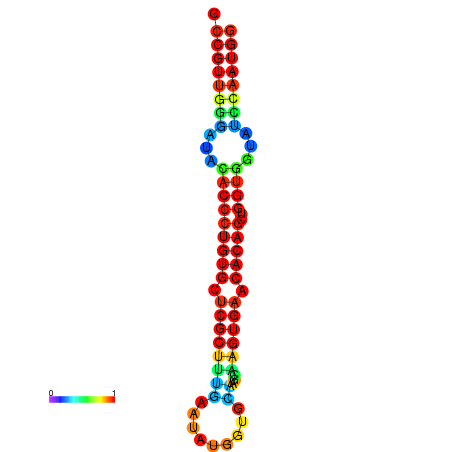 |
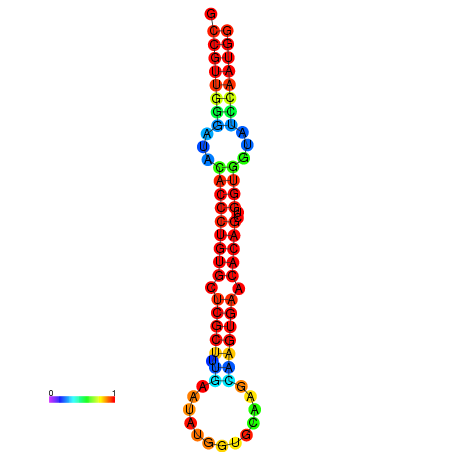 |
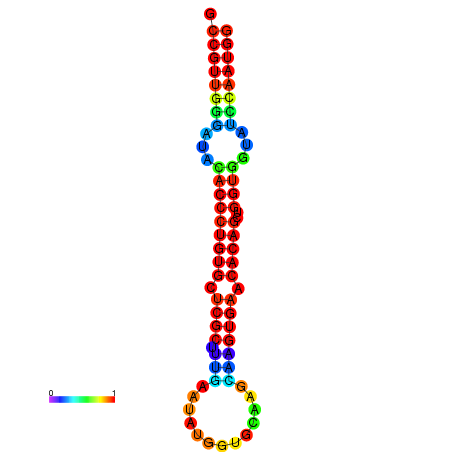 |
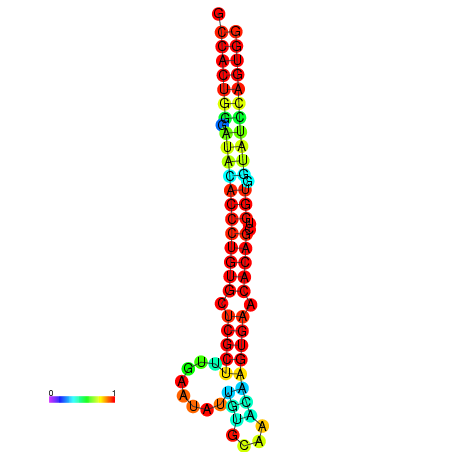
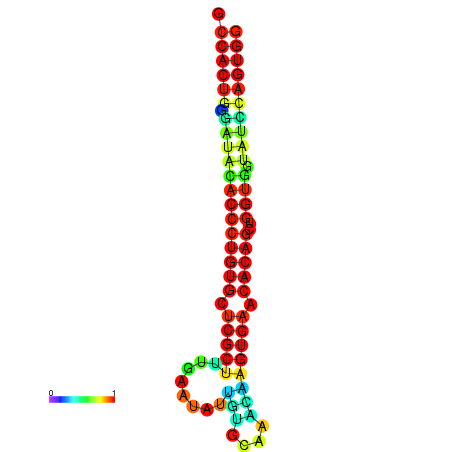
| dG=-29.9, p-value=0.009901 | dG=-29.5, p-value=0.009901 | dG=-29.5, p-value=0.009901 | dG=-29.5, p-value=0.009901 | dG=-29.5, p-value=0.009901 |
|---|---|---|---|---|
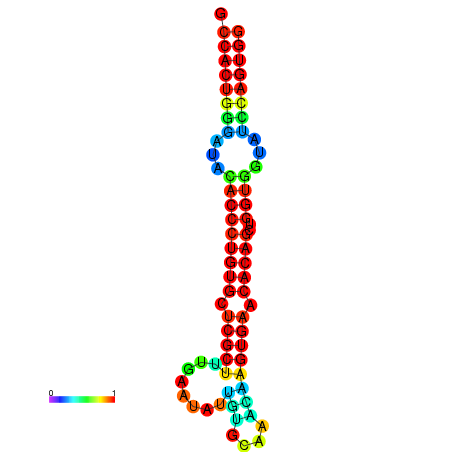 |
 |
 |
 |
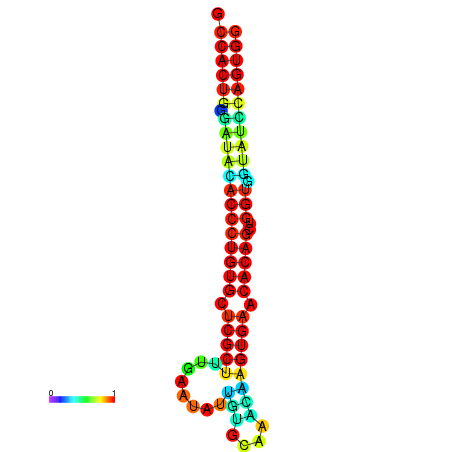 |
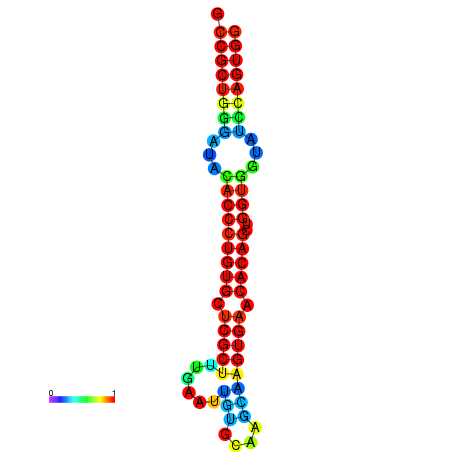
| dG=-29.5, p-value=0.009901 | dG=-29.1, p-value=0.009901 | dG=-29.1, p-value=0.009901 | dG=-29.1, p-value=0.009901 | dG=-29.1, p-value=0.009901 |
|---|---|---|---|---|
 |
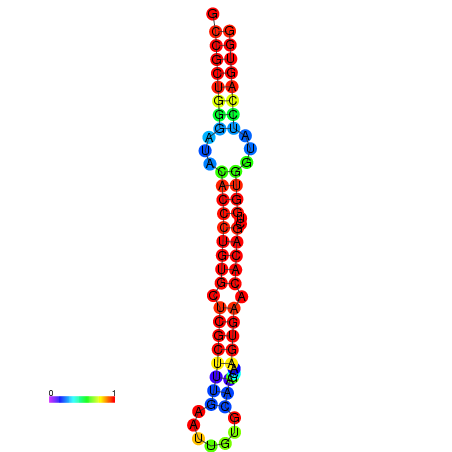 |
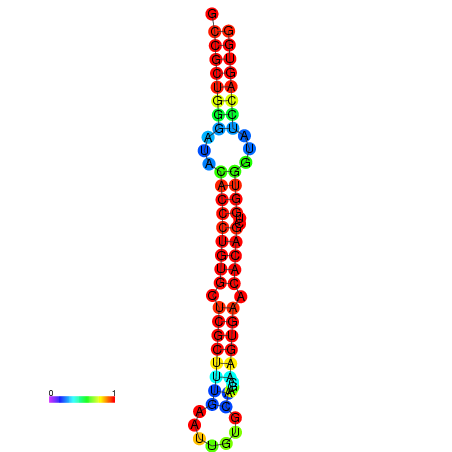 |
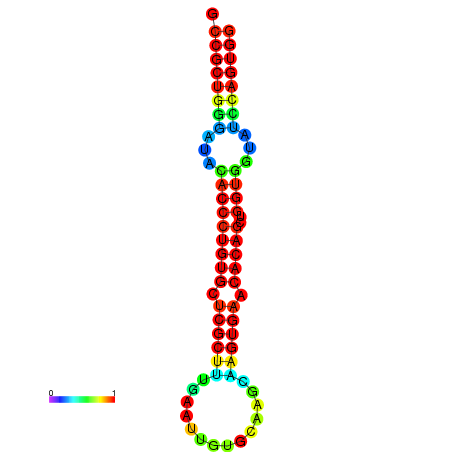 |
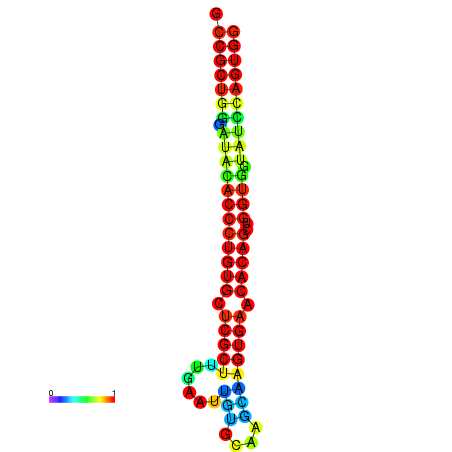 |
| dG=-26.2, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
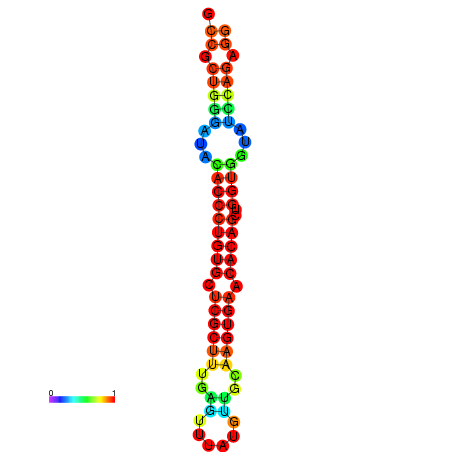 |
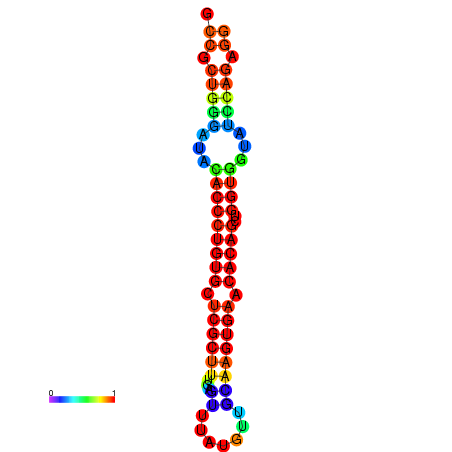 |
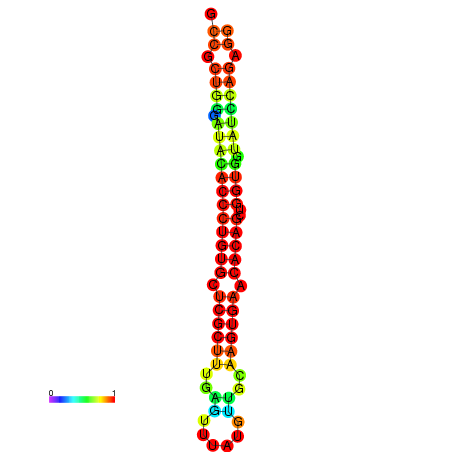 |
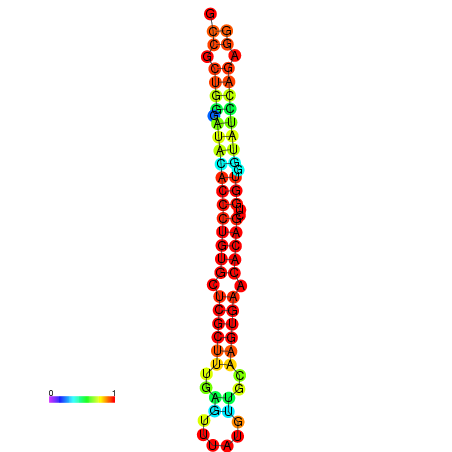 |
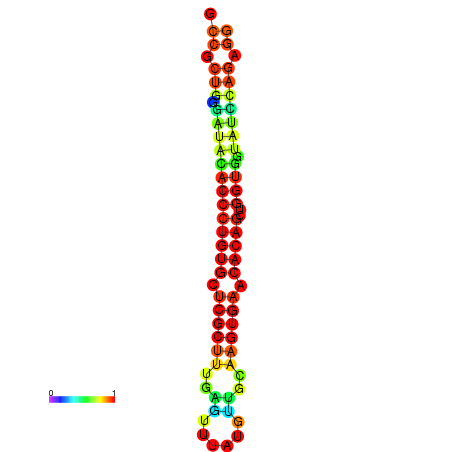 |
| dG=-31.7, p-value=0.009901 | dG=-31.7, p-value=0.009901 | dG=-31.7, p-value=0.009901 | dG=-31.7, p-value=0.009901 | dG=-31.7, p-value=0.009901 |
|---|---|---|---|---|
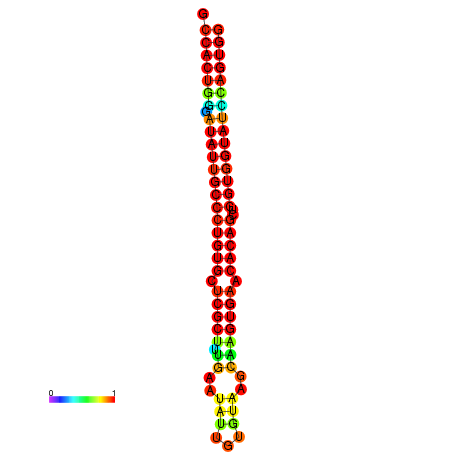 |
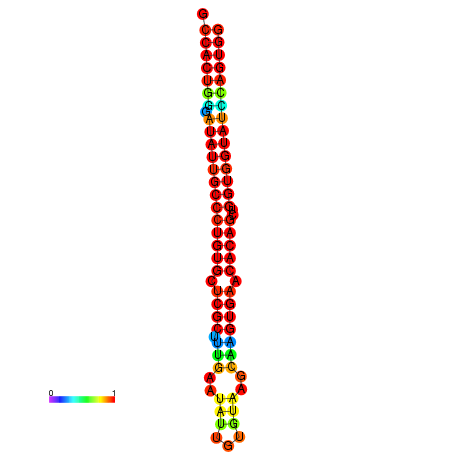 |
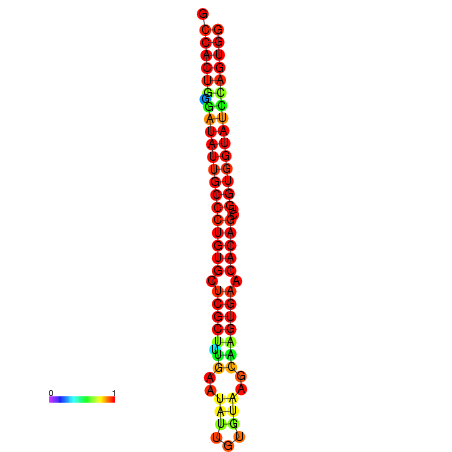 |
 |
 |
Generated: 03/07/2013 at 05:20 PM