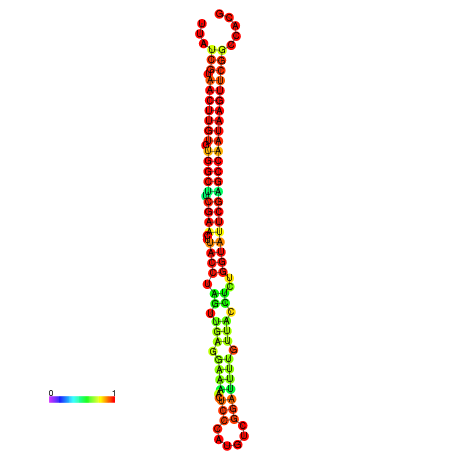
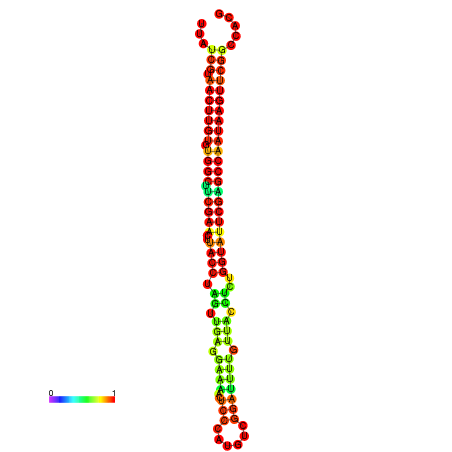
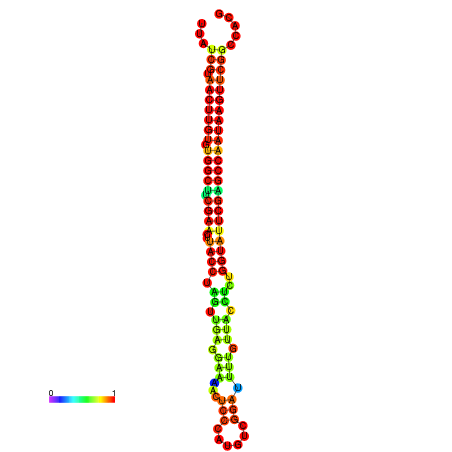
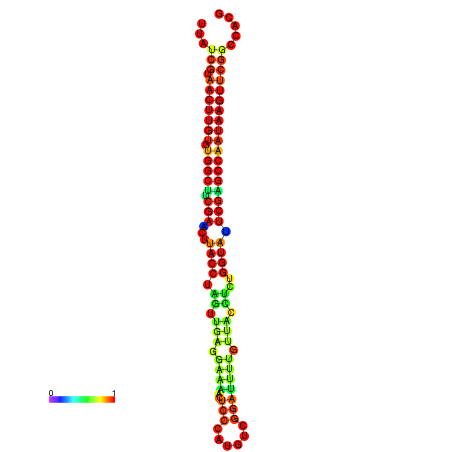
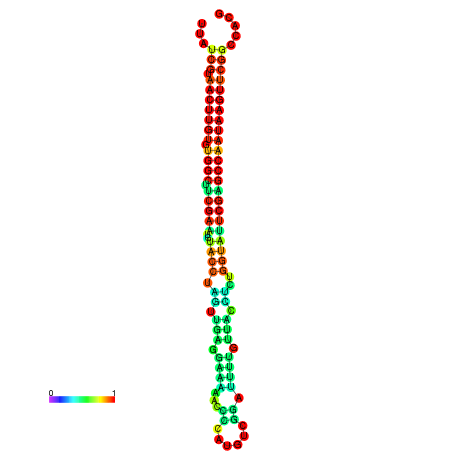
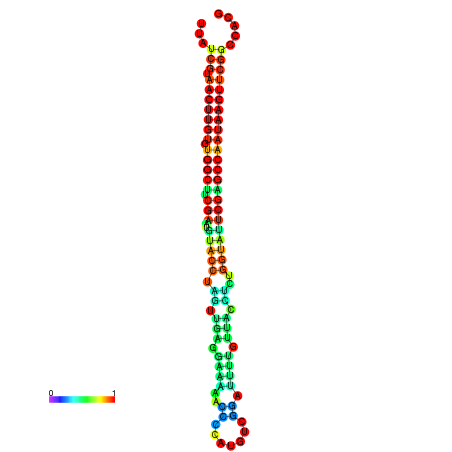
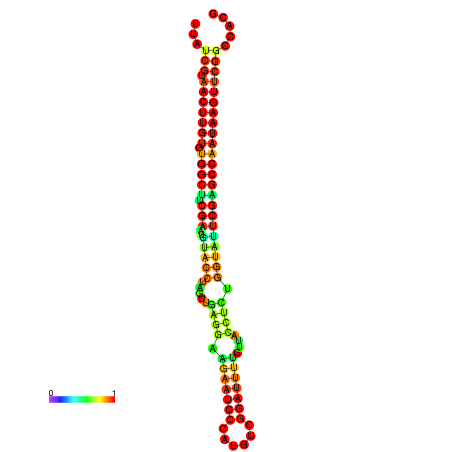
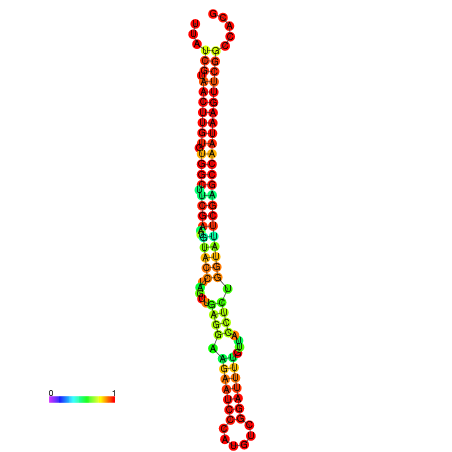
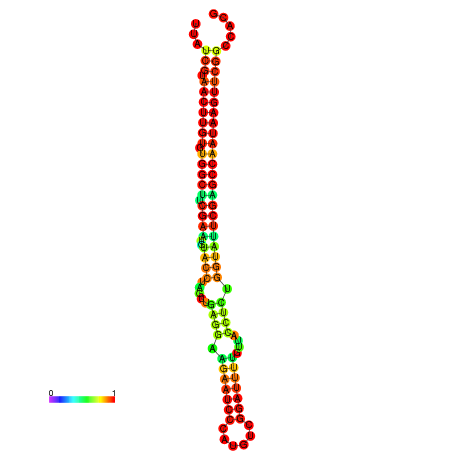
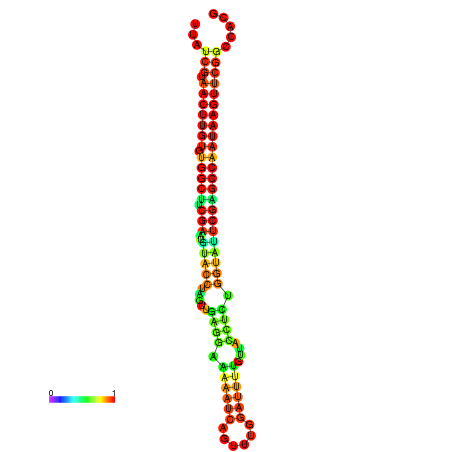
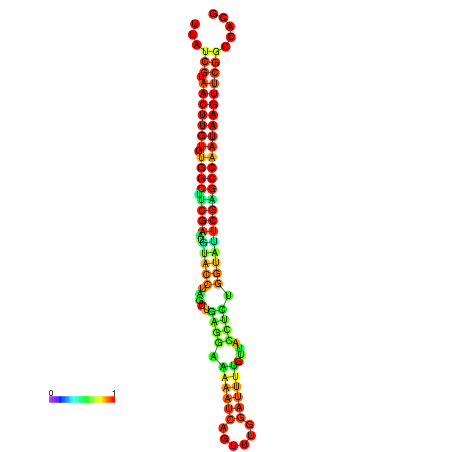
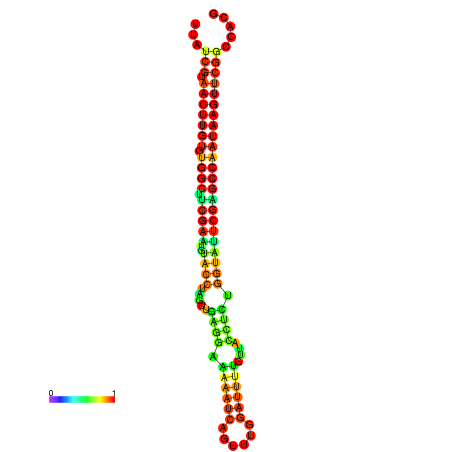
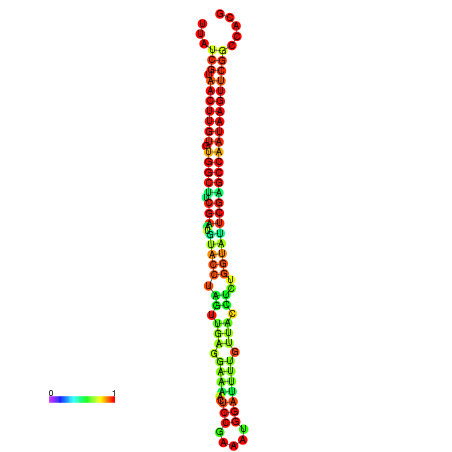
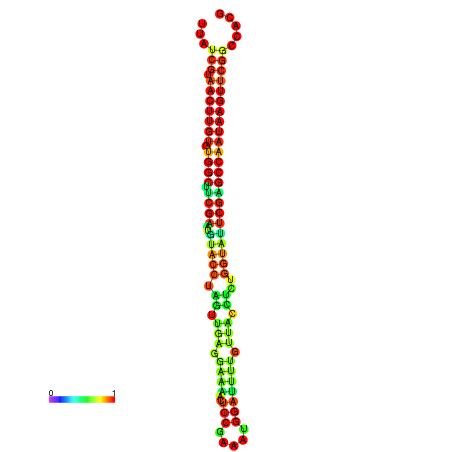
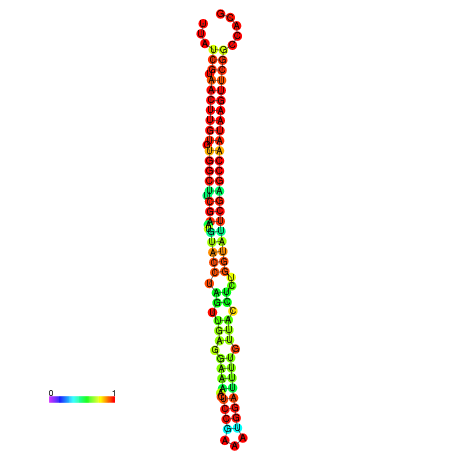
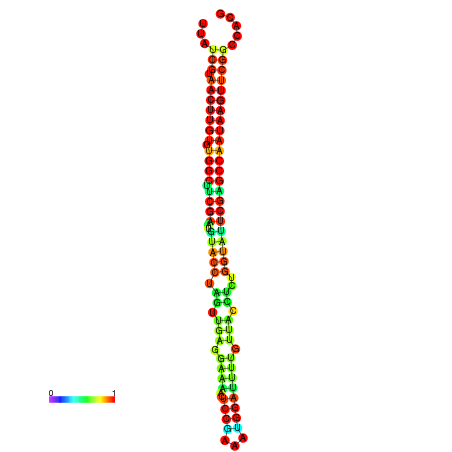
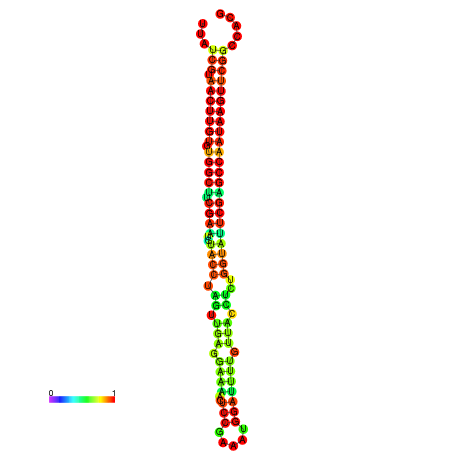
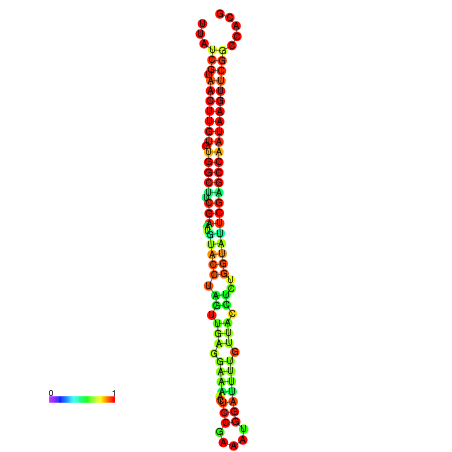
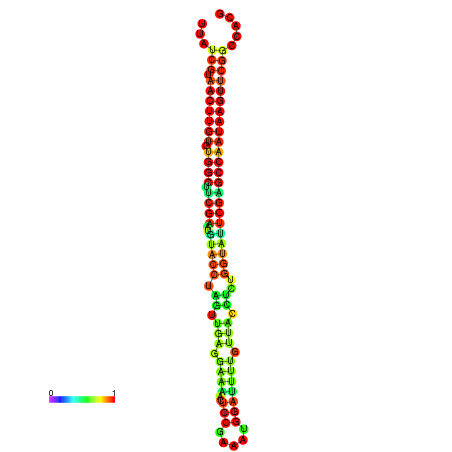
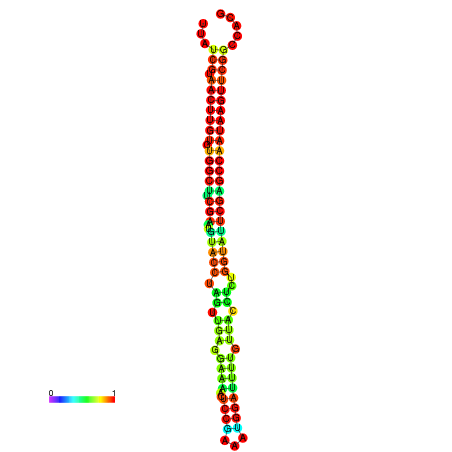
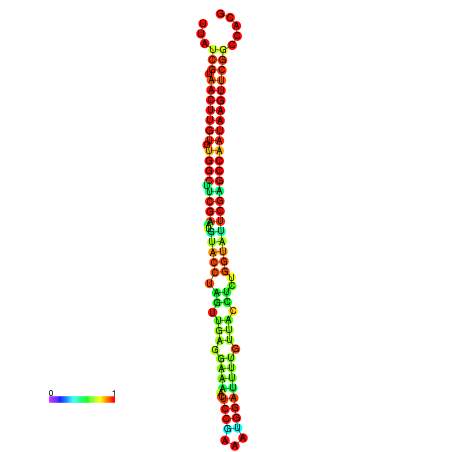
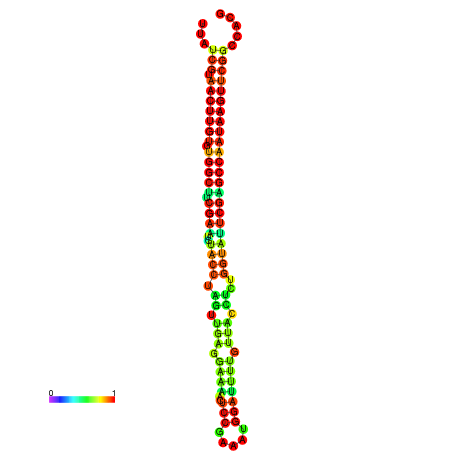
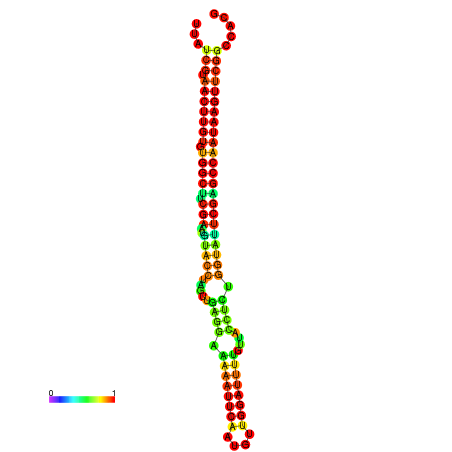
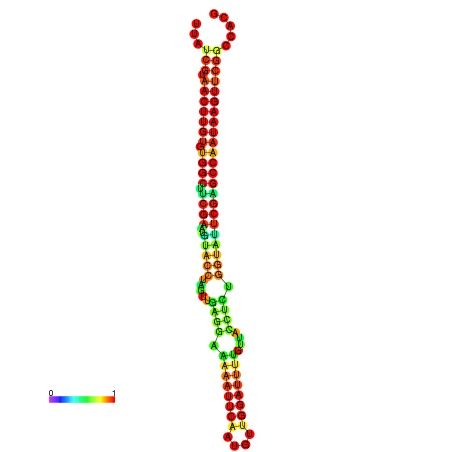
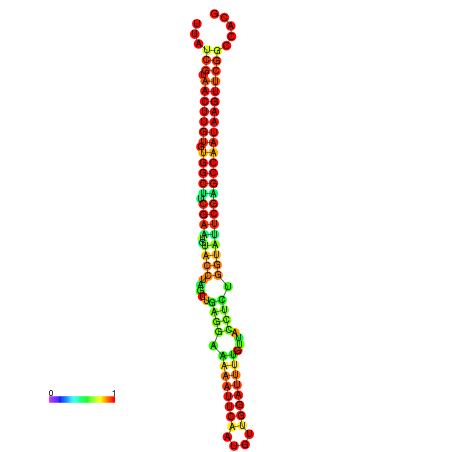
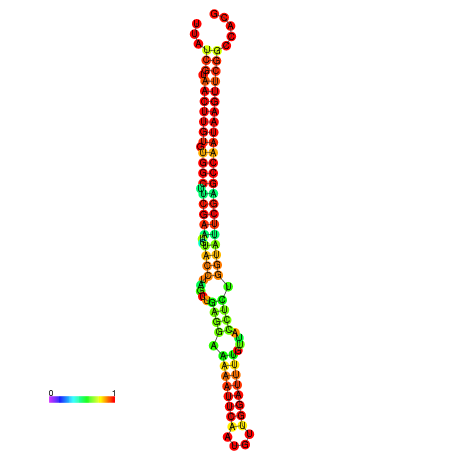
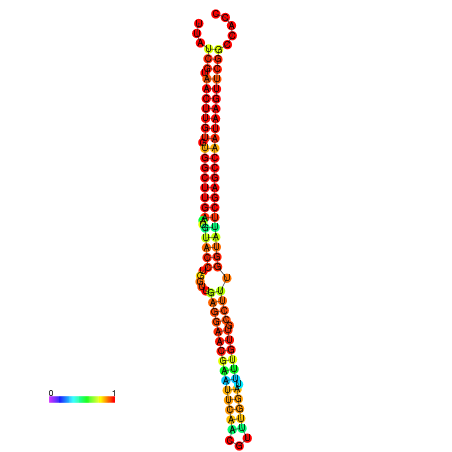
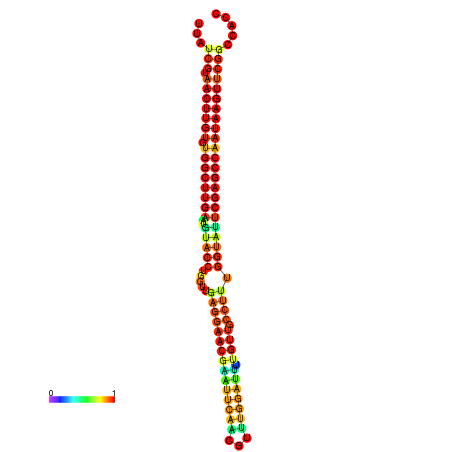
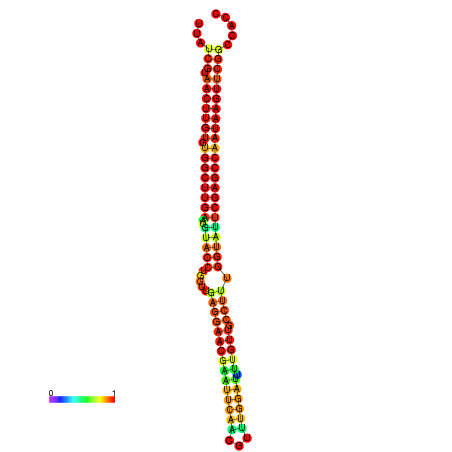
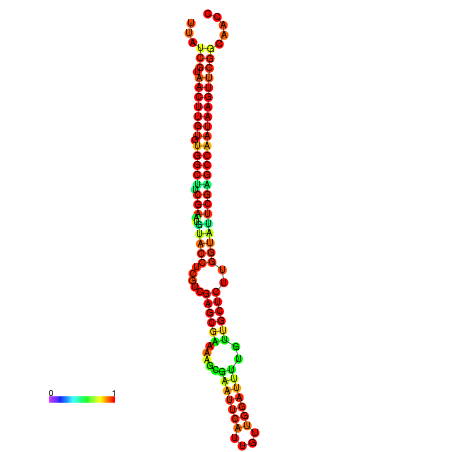
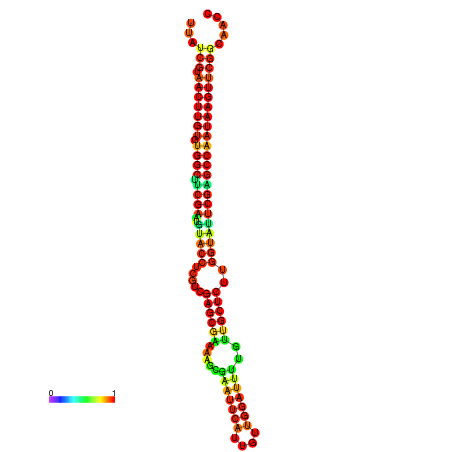
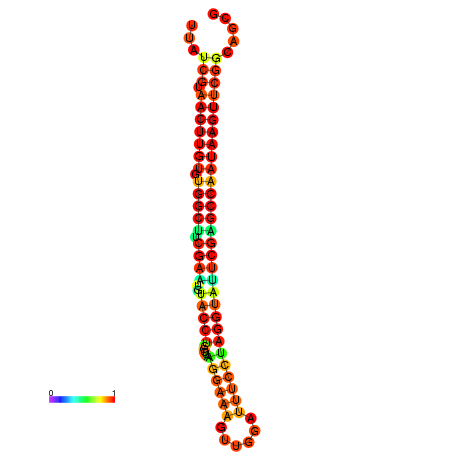
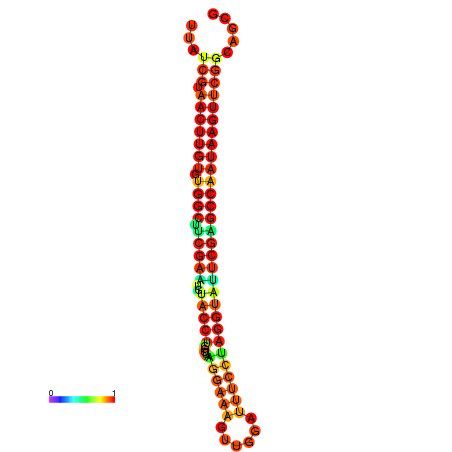
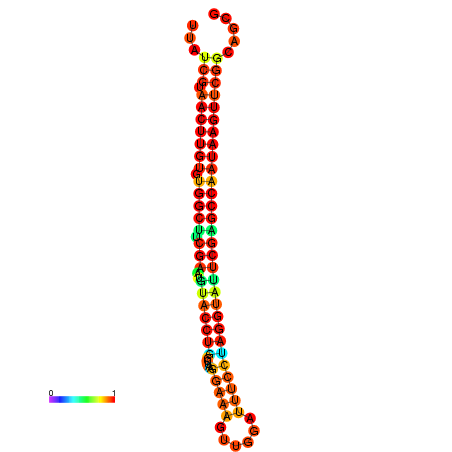
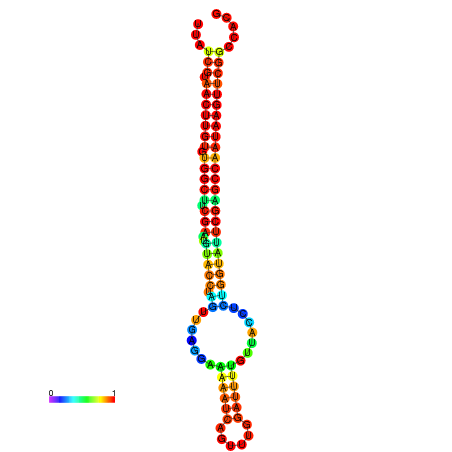| TGGTCCGCACTCTTATCGTAACTTGTGTGGCTTCGAATGTACCTAGTTGAGGAAAACTCCGA-AATGGATTTTGTTACCTCTGGTATTCGAGCCAATAAGTTCGGCCACGAAATCGCATT | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ....................................................................................TATTCGAGCCAATAAGTTCGG............... | 21 | 1 | 1790.00 | 1790 | 565 | 1225 |
| ....................................................................................TATTCGAGCCAATAAGTTCG................ | 20 | 1 | 251.00 | 251 | 17 | 234 |
| ....................................................................................TATTCGAGCCAATAAGTTCGGC.............. | 22 | 1 | 130.00 | 130 | 89 | 41 |
| .................GTAACTTGTGTGGCTTCGAATGTACC............................................................................. | 26 | 1 | 21.00 | 21 | 18 | 3 |
| ....................................................................................TATTCGAGCCAATAAGTT.................. | 18 | 1 | 12.00 | 12 | 12 | 0 |
| ....................................................................................TATTCGAGCCAATAAGTTC................. | 19 | 1 | 10.00 | 10 | 1 | 9 |
| ....................................ATGTACCTAGTTGAGGAA.................................................................. | 18 | 1 | 6.00 | 6 | 6 | 0 |
| ........................................................................TGTTACCTCTGGTATTCGAGC........................... | 21 | 1 | 5.00 | 5 | 5 | 0 |
| .......................................................................TTGTTACCTCTGGTATTCGAGC........................... | 22 | 1 | 4.00 | 4 | 4 | 0 |
| ..................TAACTTGTGTGGCTTCGAATGTACC............................................................................. | 25 | 1 | 4.00 | 4 | 4 | 0 |
| .................GTAACTTGTGTGGCTTCGA.................................................................................... | 19 | 1 | 3.00 | 3 | 3 | 0 |
| ....................................ATGTACCTAGTTGAGGAAAACT.............................................................. | 22 | 1 | 3.00 | 3 | 3 | 0 |
| .................GTAACTTGTGTGGCTTCGAAT.................................................................................. | 21 | 1 | 3.00 | 3 | 3 | 0 |
| .............................................................................................CAATAAGTTCGGCCACGA......... | 18 | 1 | 3.00 | 3 | 3 | 0 |
| .................GTAACTTGTGTGGCTTCGAATGTAC.............................................................................. | 25 | 1 | 3.00 | 3 | 3 | 0 |
| ...................................AATGTACCTAGTTGAGGAA.................................................................. | 19 | 1 | 2.00 | 2 | 2 | 0 |
| ......................TTGTGTGGCTTCGAATGTACC............................................................................. | 21 | 1 | 2.00 | 2 | 2 | 0 |
| .................................CGAATGTACCTAGTTGAGGAA.................................................................. | 21 | 1 | 2.00 | 2 | 2 | 0 |
| ..........................................................CCGA-AATGGATTTTGTTACCTCTGGT................................... | 26 | 1 | 2.00 | 2 | 2 | 0 |
| ..................................GAATGTACCTAGTTGAGGAA.................................................................. | 20 | 1 | 2.00 | 2 | 2 | 0 |
| ..................................GAATGTACCTAGTTGAGGA................................................................... | 19 | 1 | 2.00 | 2 | 2 | 0 |
| ......................................GTACCTAGTTGAGGAAAA................................................................ | 18 | 1 | 2.00 | 2 | 2 | 0 |
| ...................................AATGTACCTAGTTGAGGAAAACTCC............................................................ | 25 | 1 | 1.00 | 1 | 1 | 0 |
| ..............ATCGTAACTTGTGTGGCTTCGAAT.................................................................................. | 24 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................ACCTCTGGTATTCGAGCCAATAA..................... | 23 | 1 | 1.00 | 1 | 1 | 0 |
| ...........................................................................TACCTCTGGTATTCGAGCCAATAA..................... | 24 | 1 | 1.00 | 1 | 1 | 0 |
| ...........................................................................TACCTCTGGTATTCGAGCC.......................... | 19 | 1 | 1.00 | 1 | 1 | 0 |
| ........................GTGTGGCTTCGAATGTAC.............................................................................. | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................................................TTTGTTACCTCTGGTATTCGAGC........................... | 23 | 1 | 1.00 | 1 | 1 | 0 |
| ...........................................................CGA-AATGGATTTTGTTACCTCTGGT................................... | 25 | 1 | 1.00 | 1 | 1 | 0 |
| ..............................................................................................AATAAGTTCGGCCACGAAAT...... | 20 | 1 | 1.00 | 1 | 1 | 0 |
| ......................TTGTGTGGCTTCGAATGT................................................................................ | 18 | 1 | 1.00 | 1 | 1 | 0 |
| .................GTAACTTGTGTGGCTTCGAATGTA............................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 |
| .....................................................................................ATTCGAGCCAATAAGTTCGG............... | 20 | 1 | 1.00 | 1 | 0 | 1 |
| ...........................................................................................GCCAATAAGTTCGGCCACGAAATCGCATT | 29 | 1 | 1.00 | 1 | 1 | 0 |
| .................GTAACTTGTGTGGCTTCG..................................................................................... | 18 | 1 | 1.00 | 1 | 1 | 0 |
| .....................................TGTACCTAGTTGAGGAAAACTCC............................................................ | 23 | 1 | 1.00 | 1 | 1 | 0 |
| ........................................ACCTAGTTGAGGAAAACTCCGA-......................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 |
| .......................TGTGTGGCTTCGAATGTAC.............................................................................. | 19 | 1 | 1.00 | 1 | 1 | 0 |
| .......................TGTGTGGCTTCGAATGTACCTAGTT........................................................................ | 25 | 1 | 1.00 | 1 | 1 | 0 |
| .........................................................TCCGA-AATGGATTTTGTTACCTCTGGT................................... | 27 | 1 | 1.00 | 1 | 1 | 0 |
| .........................TGTGGCTTCGAATGTACC............................................................................. | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................................................................................CGGCCACGAAATCGCATT | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ....................................ATGTACCTAGTTGAGGAAAACTC............................................................. | 23 | 1 | 1.00 | 1 | 1 | 0 |
| ....................ACTTGTGTGGCTTCGAATGTACC............................................................................. | 23 | 1 | 1.00 | 1 | 1 | 0 |
| ..............................................................................CTCTGGTATTCGAGCCAATAAGTTCGGC.............. | 28 | 1 | 1.00 | 1 | 1 | 0 |
| ......................TTGTGTGGCTTCGAATGTACCT............................................................................ | 22 | 1 | 1.00 | 1 | 1 | 0 |
| ........................GTGTGGCTTCGAATGTACC............................................................................. | 19 | 1 | 1.00 | 1 | 1 | 0 |
| .................GTAACTTGTGTGGCTTCGAA................................................................................... | 20 | 1 | 1.00 | 1 | 1 | 0 |
| ...........................................TAGTTGAGGAAAACTCCGA-AATGGATT................................................. | 27 | 1 | 1.00 | 1 | 1 | 0 |
| .................GTAACTTGTGTGGCTTCGAATGTACCT............................................................................ | 27 | 1 | 1.00 | 1 | 1 | 0 |
| .....................................................................................ATTCGAGCCAATAAGTTCGGCCACGA......... | 26 | 1 | 1.00 | 1 | 1 | 0 |
| ..........................................................CCGA-AATGGATTTTGTTACC......................................... | 20 | 1 | 1.00 | 1 | 1 | 0 |
| ....................ACTTGTGTGGCTTCGAATGTACCT............................................................................ | 24 | 1 | 1.00 | 1 | 1 | 0 |
| ........................GTGTGGCTTCGAATGTACCTAGTTGA...................................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................ACCTCTGGTATTCGAGCCAATA...................... | 22 | 1 | 1.00 | 1 | 1 | 0 |
| ........................................................................TGTTACCTCTGGTATTCGAGCCA......................... | 23 | 1 | 1.00 | 1 | 1 | 0 |
| ...............................................TGAGGAAAACTCCGA-AATGGATTTT............................................... | 25 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................GTACCTAGTTGAGGAAAACT.............................................................. | 20 | 1 | 1.00 | 1 | 1 | 0 |
| ........................................................................TGTTACCTCTGGTATTCGAGCCAATAA..................... | 27 | 1 | 1.00 | 1 | 1 | 0 |
| ..........................GTGGCTTCGAATGTACCTAGTTGAGG.................................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 |
| .........................................................................................GAGCCAATAAGTTCGGCCACGAAATCGC... | 28 | 1 | 1.00 | 1 | 1 | 0 |
| ...................................AATGTACCTAGTTGAGGA................................................................... | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ...................................................................................GTATTCGAGCCAATAAGTTCGG............... | 22 | 1 | 1.00 | 1 | 1 | 0 |
| ...........................................................CGA-AATGGATTTTGTTACCTCT...................................... | 22 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................GA-AATGGATTTTGTTACCTC....................................... | 20 | 1 | 1.00 | 1 | 1 | 0 |
| ..........................................................CCGA-AATGGATTTTGTTAC.......................................... | 19 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................GA-AATGGATTTTGTTACCTCTGG.................................... | 23 | 1 | 1.00 | 1 | 1 | 0 |
| .................................CGAATGTACCTAGTTGAGGA................................................................... | 20 | 1 | 1.00 | 1 | 1 | 0 |
| ........................................................CTCCGA-AATGGATTTTGTTACCTCTGGT................................... | 28 | 1 | 1.00 | 1 | 1 | 0 |
| ...................AACTTGTGTGGCTTCGAAT.................................................................................. | 19 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................ACCTCTGGTATTCGAGCCAATAAGT................... | 25 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................GTACCTAGTTGAGGAAAACTCC............................................................ | 22 | 1 | 1.00 | 1 | 1 | 0 |
| ..............................CTTCGAATGTACCTAGTTGAG..................................................................... | 21 | 1 | 1.00 | 1 | 1 | 0 |
| .....................................TGTACCTAGTTGAGGAAAACT.............................................................. | 21 | 1 | 1.00 | 1 | 1 | 0 |
| ..................................GAATGTACCTAGTTGAGGAAAACT.............................................................. | 24 | 1 | 1.00 | 1 | 1 | 0 |
| ................................................................................CTGGTATTCGAGCCAATAAGTTCGGC.............. | 26 | 1 | 1.00 | 1 | 1 | 0 |