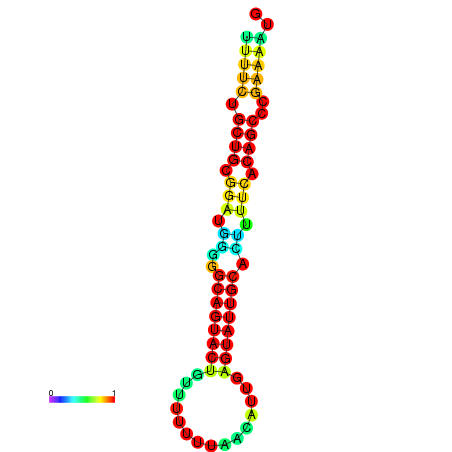
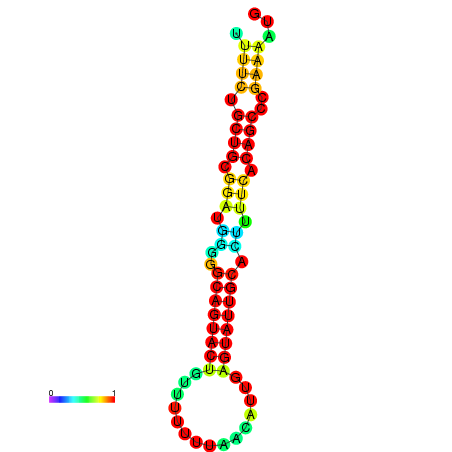
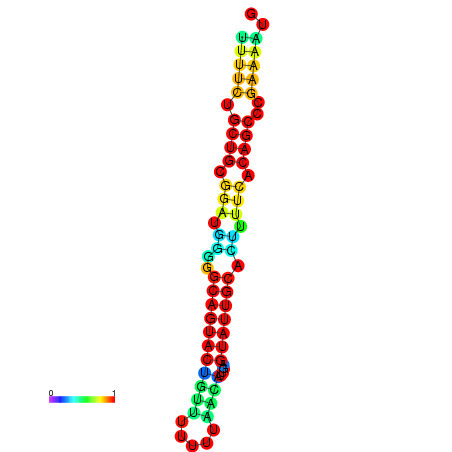
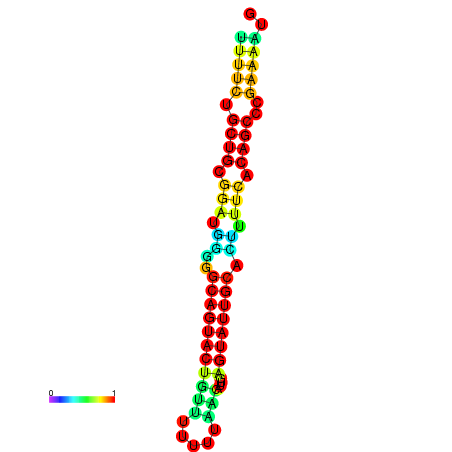
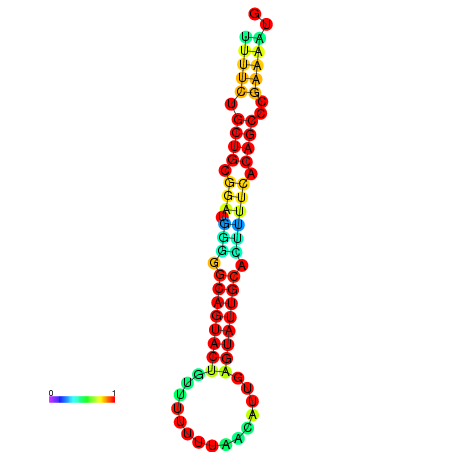
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16471680-16471780 - | TCGTTCT--AGCCAACAA---TTTTCTGCTGCGGATGGGGGCAGTACTG--------------------------TTTTTTTAACATT--GAGTATTGCACTTTTCACAGCCCG-----AAAATGTTAGCAACTGC-CTT |
| droSim1 | chr2R:15132971-15133071 - | TCGATCT--AGCCAACAA---TTTTCTGCTGCGCATTGGGGCACTACTG--------------------------TTTTTTTTAAATT--GAGTATTGCACTTTTCACAGCCTT-----AAAATGTTAGCAACTGC-CTA |
| droSec1 | super_1:14025885-14025985 - | TCGATCT--AGCCAACAA---TTTTCTGCTGCGCATTGGGGCACTACTG--------------------------TTTTTTTTTAATT--GAGTATTGCACTTTTCACAGCCTT-----AAAATGTTAGCAACTGC-CTA |
| droYak2 | chr2R:12724540-12724640 + | TCTACGT--AGCCAACAA---TTTTCGGCTGTGGATTGGGCTAGTACTG--------------------------TGATTTTAGCATT--AAGTATTGCACATTCCCCAGCCTG-----AAAGTGTTAGTAACTGC-CAA |
| droEre2 | scaffold_4845:10618281-10618381 - | TCGATGT--AGCCAACAA---TTTTCGGCTGTGGAGTGGGCTAGTACTG--------------------------TGATTTTAACATT--GAGTATTGCACATTCCCCAGCCTG-----AAAGTGTTTGCATTTGC-CAA |
| droAna3 | scaffold_13266:4132526-4132623 + | TTT--GT--AGTCAGCAT---TTCCAGACTGTGGTTTGGGCAGATACTA--------------------------CATTTT--AGGAT--CAGTATTGCACATTCCCCAGCCTG-----GAAACGCTTGCGACAGA-CCT |
| dp4 | Unknown | -------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer1 | super_4:2566453-2566554 + | TTCGTGTTCGGTGGATAC---CTTCAAGCTGGGGGATGGG-CAATACTG--------------------------TGTTGTATTTGAA--CAGTATTGCACACCCACTGGCCTG-----AAAGTGCCTACTGCTGG-GTT |
| droWil1 | scaffold_180700:3452993-3453102 + | TTAATTATTTGTCCAAATTTAGTTCTGGCTGCGGTTTGTGCAAATTACT---T----------------------ATTTT--TTTGTA--AAGTATTGCACAACCTACAGCCTGGCACTAAATGGAGAAAAATCAT-CAA |
| droVir3 | scaffold_12875:13669100-13669187 - | T---------------------TTTCGGCTGTGGTCAGTGTCAATTCTT--------------------------TTTTTTTATTTTTACAAGTATTGCACTTTTCACGGCCGG-----AAAAATTGAGATTCCATTTAA |
| droMoj3 | scaffold_6496:9090655-9090750 - | TTT--TT--TGCATTCGT---TTTCCGGTTGTGAATTGTGTCAATTCTT--------------------------TTTATTTTAC-----AAGTATTGCACTTTTCACGGCCGG-----AAAAAGGATAAAATATT-TAA |
| droGri2 | scaffold_15245:17564118-17564229 + | TCATTAT--CCCGAACAA---CATCATTCCGCGCTCGGCTTTAGAATTTGTTCCTTATTTTGTGAGGATAATTTACAATTC--GAAGA--TAATAT-----------TGGC-AG-----GATATG-TTACAGTTCC-TCT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16471680-16471780 - | TCGTTC--TAGCCA-A--CAATTTTCTGCTGCGGATGGGGGCAGTA-CTGTTTTTTTAACATT--GAGTATTGCACTTTTCACAGCCCG-----AAAATGTTAGCAACT-GCCTT |
| droSim1 | chr2R:15132971-15133071 - | TCGATC--TAGCCA-A--CAATTTTCTGCTGCGCATTGGGGCACTA-CTGTTTTTTTTAAATT--GAGTATTGCACTTTTCACAGCCTT-----AAAATGTTAGCAACT-GCCTA |
| droSec1 | super_1:14025885-14025985 - | TCGATC--TAGCCA-A--CAATTTTCTGCTGCGCATTGGGGCACTA-CTGTTTTTTTTTAATT--GAGTATTGCACTTTTCACAGCCTT-----AAAATGTTAGCAACT-GCCTA |
| droYak2 | chr2R:12724540-12724640 + | TCTACG--TAGCCA-A--CAATTTTCGGCTGTGGATTGGGCTAGTA-CTGTGATTTTAGCATT--AAGTATTGCACATTCCCCAGCCTG-----AAAGTGTTAGTAACT-GCCAA |
| droEre2 | scaffold_4845:10618281-10618381 - | TCGATG--TAGCCA-A--CAATTTTCGGCTGTGGAGTGGGCTAGTA-CTGTGATTTTAACATT--GAGTATTGCACATTCCCCAGCCTG-----AAAGTGTTTGCATTT-GCCAA |
| droAna3 | scaffold_13266:4132526-4132623 + | TT--TG--TAGTCA-G--CATTTCCAGACTGTGGTTTGGGCAGATA-CTACATTTT--AGGAT--CAGTATTGCACATTCCCCAGCCTG-----GAAACGCTTGCGACA-GACCT |
| droPer1 | super_4:2566453-2566554 + | TTCGTGTTCGGTGG-A--TACCTTCAAGCTGGGGGATGGG-CAATA-CTGTGTTGTATTTGAA--CAGTATTGCACACCCACTGGCCTG-----AAAGTGCCTACTGCT-GGGTT |
| droWil1 | scaffold_180700:3452993-3453102 + | TTAATTATTTGTCCAAATTTAGTTCTGGCTGCGGTTTGTGCAAATTACTTATTT--TTTTGTA--AAGTATTGCACAACCTACAGCCTGGCACTAAATGGAGAAAAATC-ATCAA |
| droVir3 | scaffold_12875:13669100-13669187 - | T---------------------TTTCGGCTGTGGTCAGTGTCAATT-CTTTTTTTTTATTTTTACAAGTATTGCACTTTTCACGGCCGG-----AAAAATTGAGATTCCATTTAA |
| droMoj3 | scaffold_6496:9090655-9090750 - | TTT--T--TTGCAT-T--CGTTTTCCGGTTGTGAATTGTGTCAATT-CTTTTTATTTTAC-----AAGTATTGCACTTTTCACGGCCGG-----AAAAAGGATAAAATA-TTTAA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-20.3, p-value=0.009901 | dG=-20.0, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
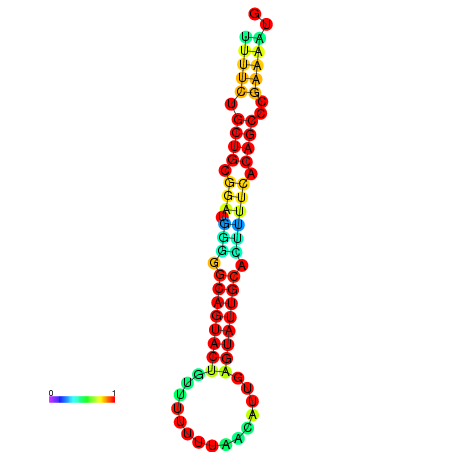 |
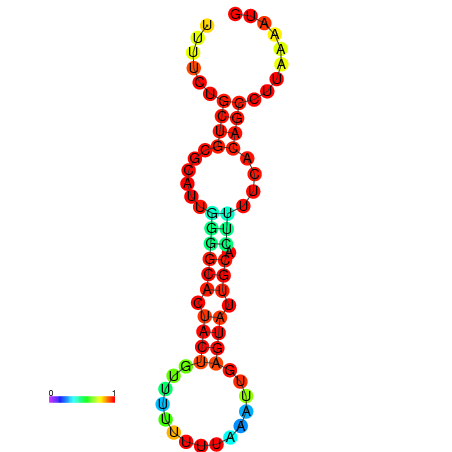
| dG=-10.7, p-value=0.009901 | dG=-10.7, p-value=0.009901 | dG=-10.7, p-value=0.009901 | dG=-10.7, p-value=0.009901 | dG=-10.2, p-value=0.009901 |
|---|---|---|---|---|
 |
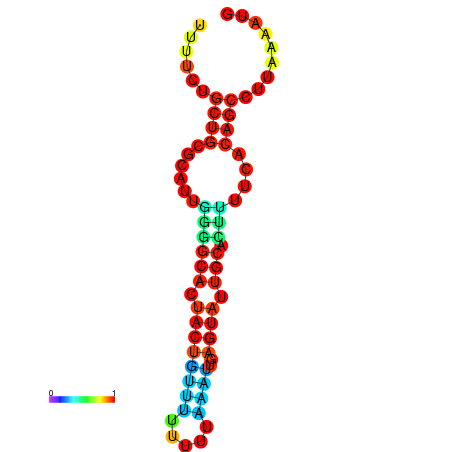 |
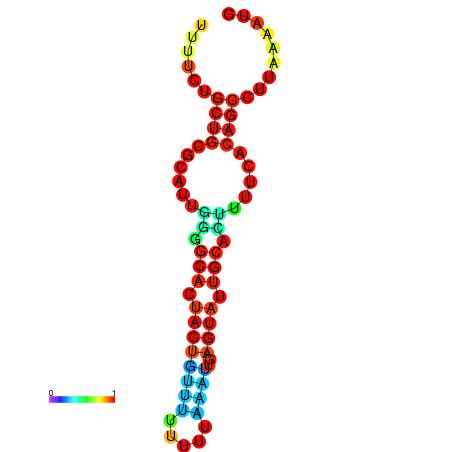 |
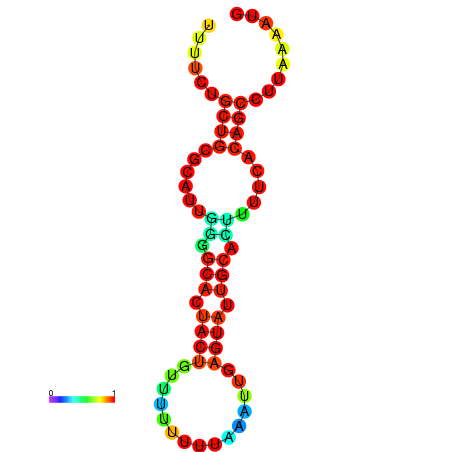 |
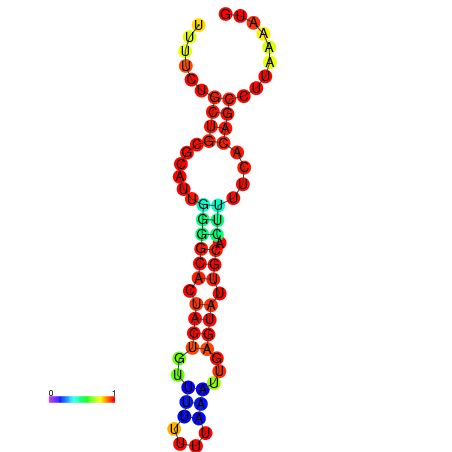 |
| dG=-10.7, p-value=0.009901 | dG=-10.7, p-value=0.009901 | dG=-10.4, p-value=0.009901 | dG=-10.4, p-value=0.009901 | dG=-9.8, p-value=0.009901 |
|---|---|---|---|---|
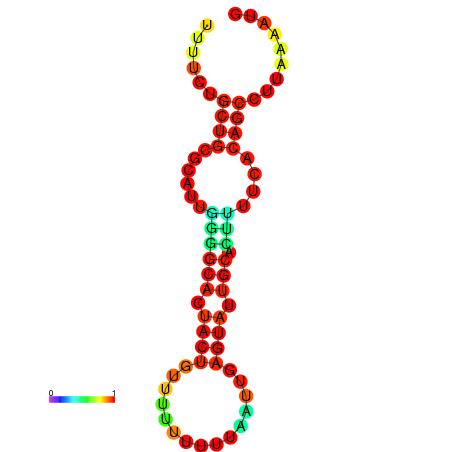 |
 |
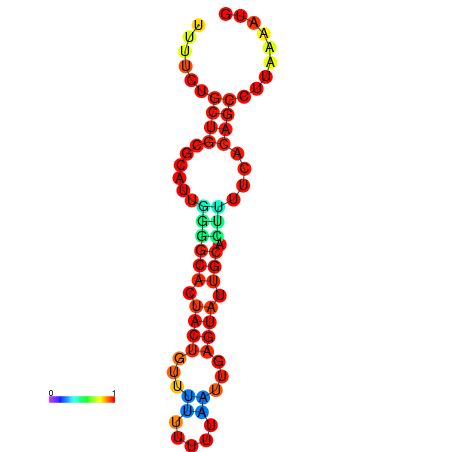 |
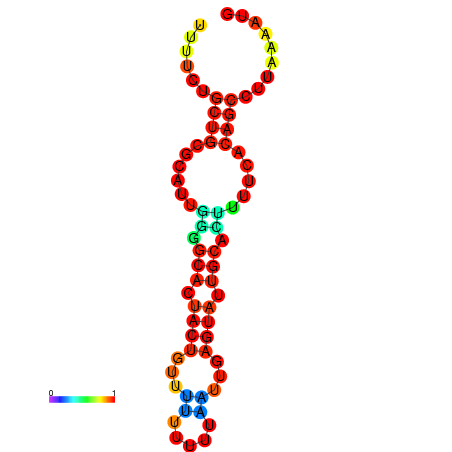 |
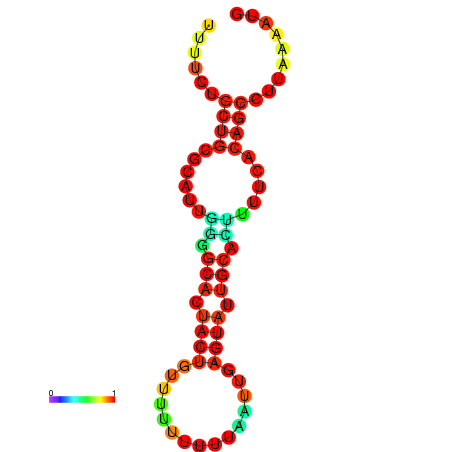
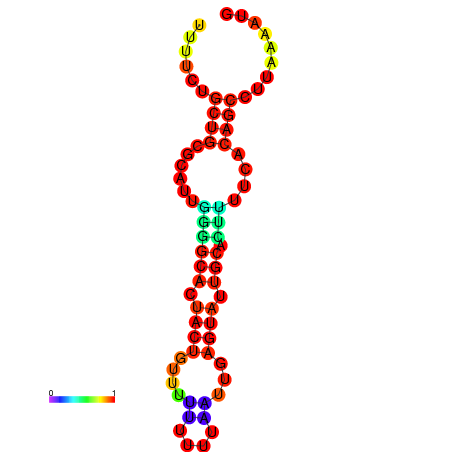 |
| dG=-23.9, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.7, p-value=0.009901 | dG=-23.7, p-value=0.009901 | dG=-23.3, p-value=0.009901 |
|---|---|---|---|---|
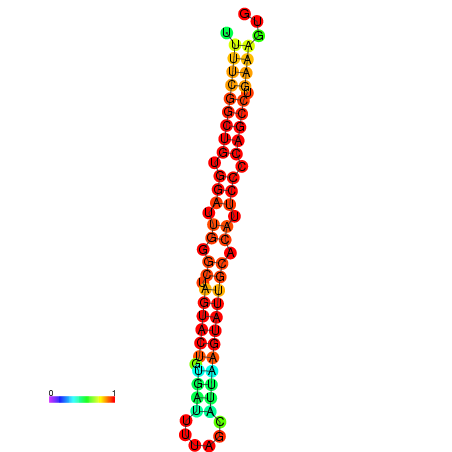 |
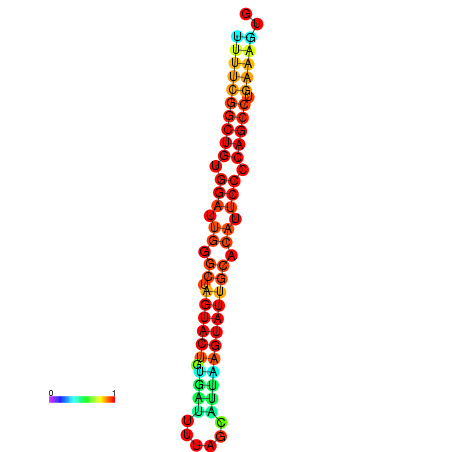 |
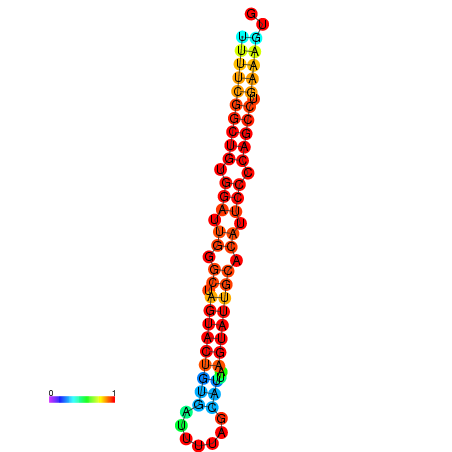 |
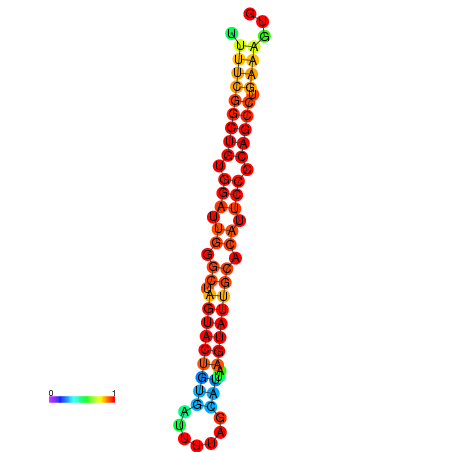 |
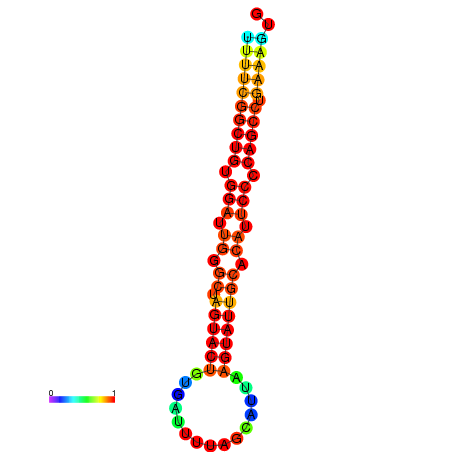 |
| dG=-26.7, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.4, p-value=0.009901 |
|---|---|---|---|---|
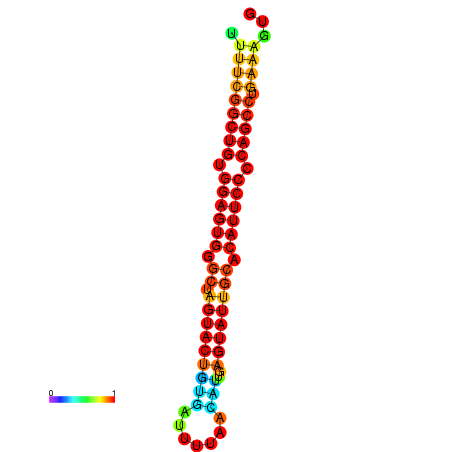 |
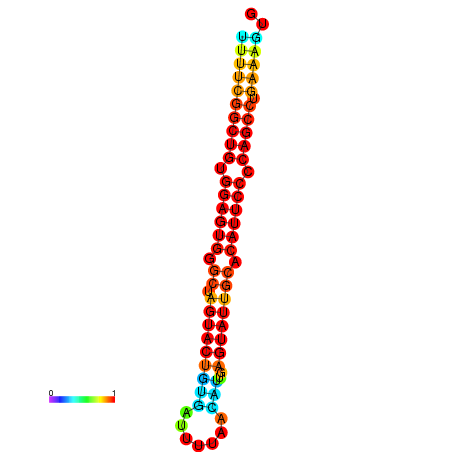 |
 |
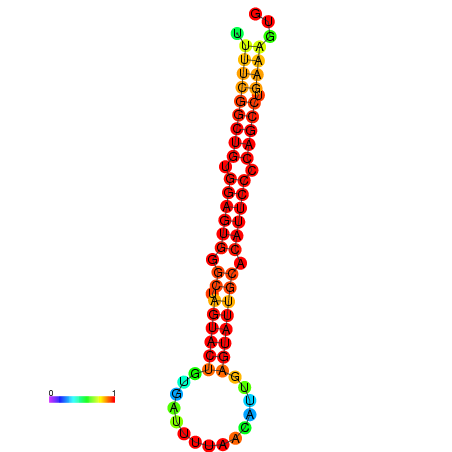 |
 |
| dG=-21.8, p-value=0.009901 | dG=-21.8, p-value=0.009901 | dG=-21.6, p-value=0.009901 | dG=-21.6, p-value=0.009901 | dG=-21.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
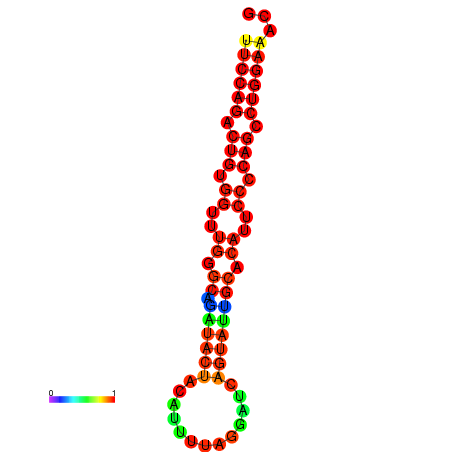 |
No secondary structure predictions returned
| dG=-30.1, p-value=0.009901 | dG=-29.4, p-value=0.009901 | dG=-29.4, p-value=0.009901 | dG=-29.2, p-value=0.009901 |
|---|---|---|---|
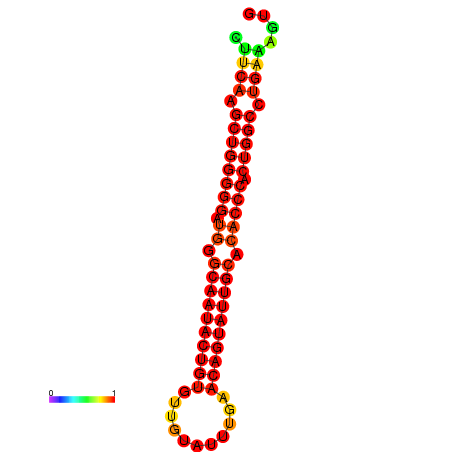 |
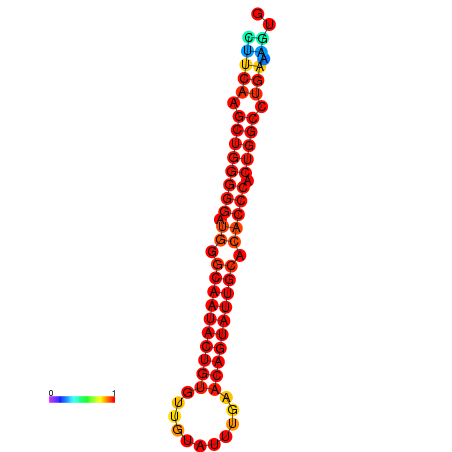 |
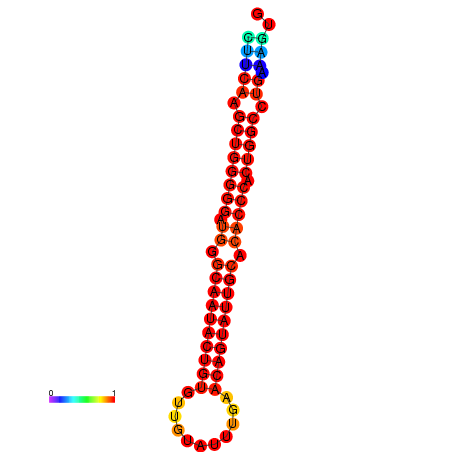 |
 |
| dG=-23.3, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-23.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
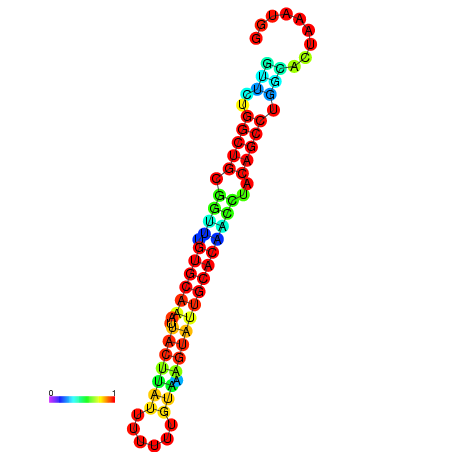 |
| dG=-24.3, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.4, p-value=0.009901 | dG=-23.3, p-value=0.009901 |
|---|---|---|---|
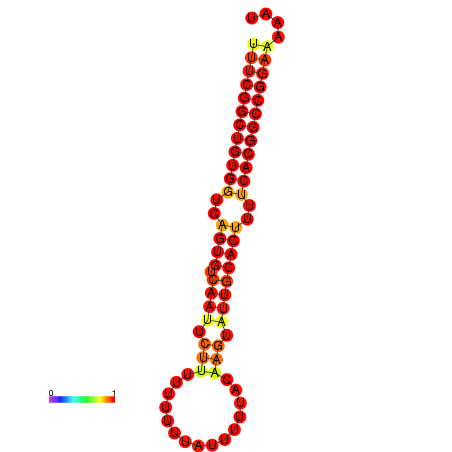 |
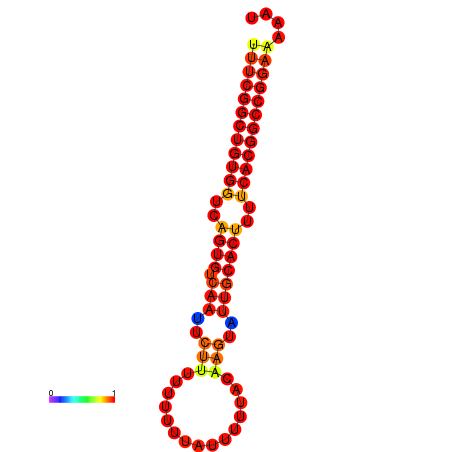 |
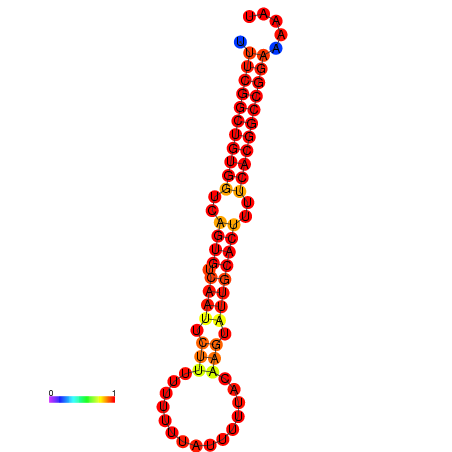 |
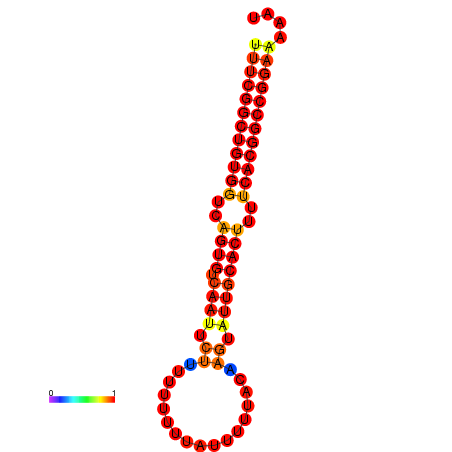 |
| dG=-28.7, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.7, p-value=0.009901 |
|---|---|---|---|
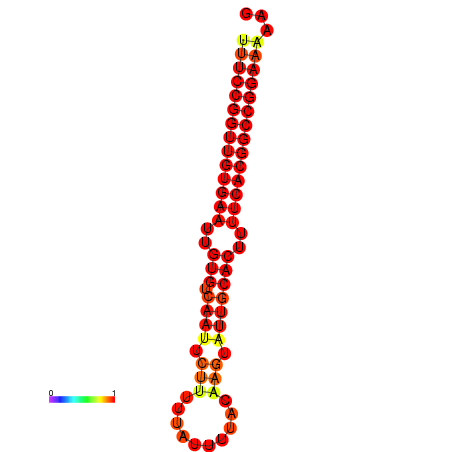 |
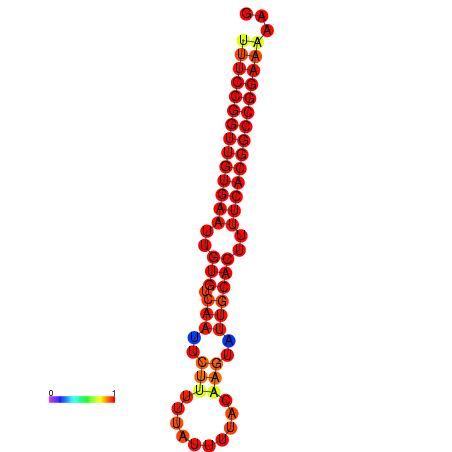 |
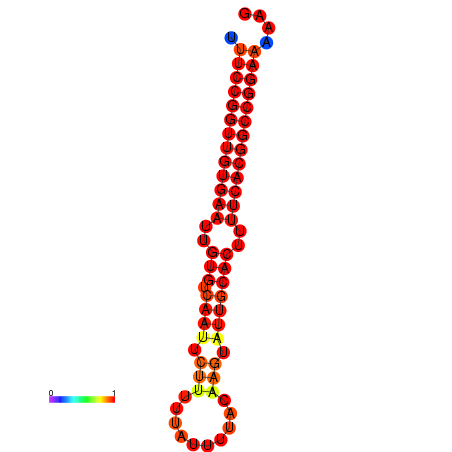 |
 |
| dG=-13.0, p-value=0.009901 | dG=-13.0, p-value=0.009901 | dG=-13.0, p-value=0.009901 | dG=-12.8, p-value=0.009901 | dG=-12.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
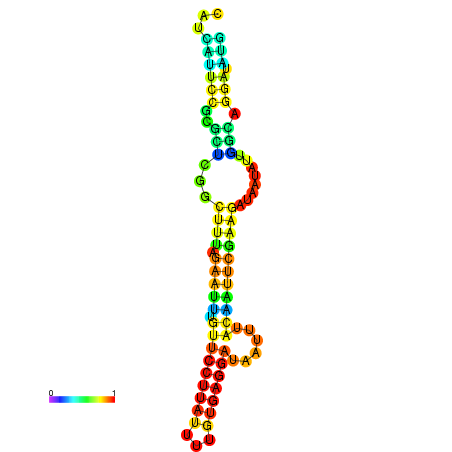 |
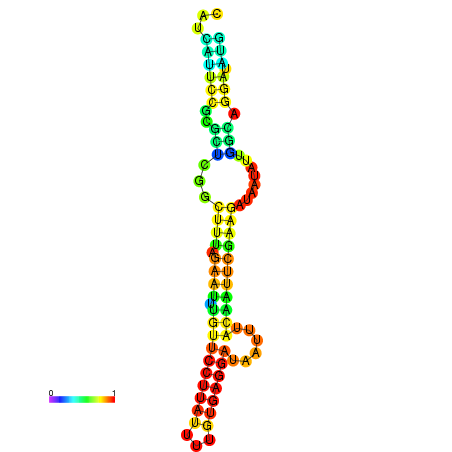 |
Generated: 03/07/2013 at 05:17 PM