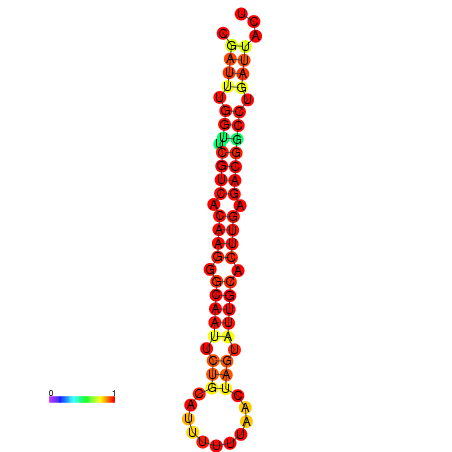
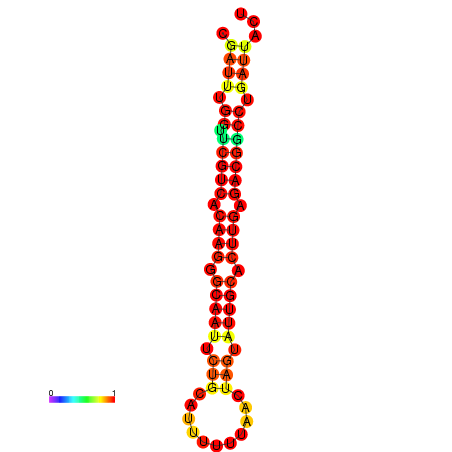
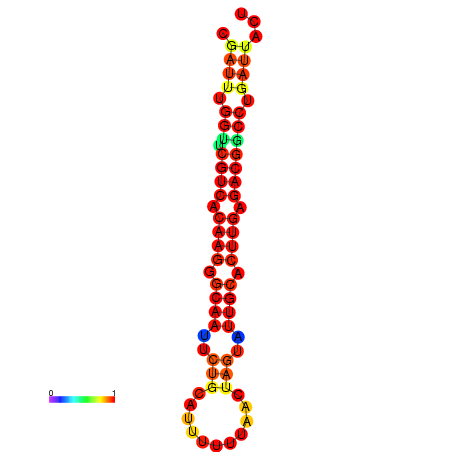
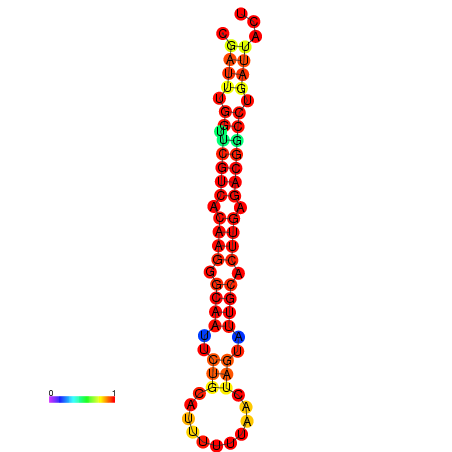
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16471546-16471641 - | AATTAAGTT-TGAAGCGATTTGGTTCGTCACAAGGGCAATTCTGCATT--TTTTAACTAGTATTGCACTTGAGACGGCCTGATTA--CTTTA-----AAACGAATC |
| droSim1 | chr2R:15132834-15132929 - | AATTAAGTT-TAAAGCGTTTTGGTTCGTCACAAGGGCAATTCTGCATT--TTTAAACTTGTATTGCACTTGAGACGGCCGAATTA--CTTTA-----AAAAGAATC |
| droSec1 | super_1:14025748-14025843 - | AATTAAGTT-TAAAGCGTTTTGGTTCGTCACAAGGGCAATTCTGCATT--TTTAAACTTGTATTGCACTTGAGACGGCCGAATTA--CTTTA-----AAAAGAATC |
| droYak2 | chr2R:12724685-12724781 + | AATTGCGTT-TGAAGCGCTTCGGTTCGTCCCAGGAGCAATTCTGCACTA-TTAAAGCTAGTATTGCACTTGAGACGGCCGGATTA--CTTTT-----AAACGAATC |
| droEre2 | scaffold_4845:10618147-10618242 - | AATTAAGTT-TGAAGCGCTTCGGTTCGTACCAGGAGCAATTCTGCATT--TTTAAGCTAGTATTGCACTTGAGACGGCCGGATTA--CTTTT-----AAACAAAAC |
| droAna3 | scaffold_13266:4132676-4132765 + | TCTACAAT--------TTTCCGGTGCGTCTCTAGTGCAACTCTTGGTTT-TTTATACAAGTATTGCACTTGAGACGGCCGAAATA--TAGTG-----GATCTAAGC |
| dp4 | chr3:8679756-8679828 + | G---------------------------------GGCAATACTGTGTTGTATTTCACCAGTATTGCACACCCACTGGCCTGAAAGTGCCTACTGCTGGGTTCAACA |
| droPer1 | gnl|ti|732455623:23-94 + | G---------------------------------GGCAATACTGTGTTGTATTTGAACAGTATTGCACACCCACTGGCCTGAAAGTGCCTACTGCTGGGTTCAA-- |
| droWil1 | scaffold_180700:3453141-3453235 + | CATAAACTCGAAAATTTTTTAGGTTCGTTTAGTCTGCAAATT-GTAAT--ATACGAAAATAATTGCACACTTCACGGCCGGAAAA--GTTCA-----AGTTAAACA |
| droVir3 | scaffold_12875:13669041-13669063 - | AGCTATATT-T-----------------------------------------------------------------------------TTCA-----GGCCGTGTC |
| droMoj3 | scaffold_6496:9090603-9090622 - | GTCTGCAAT-TTAAGCTTCTT------------------------------------------------------------------------------------- |
| droGri2 | scaffold_15245:17564268-17564358 + | AAGCAAATC-TTCAGTTTTTCGGCTGTGGTTAGTGTCAATTCTTTTT---ATTTGACAAGCATTGCACTCTTCACGGCCGGATAA--AT---------GAGGGTCT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16471546-16471641 - | AATTAAGTTT-GAAGCGATTTGGTTCGTCACAAGGGCAATTCTGCATT--TTTTAACTAGTATTGCACTTGAGACGGCCTGATTA--CTTTAAAACGAA-----TC |
| droSim1 | chr2R:15132834-15132929 - | AATTAAGTTT-AAAGCGTTTTGGTTCGTCACAAGGGCAATTCTGCATT--TTTAAACTTGTATTGCACTTGAGACGGCCGAATTA--CTTTAAAAAGAA-----TC |
| droSec1 | super_1:14025748-14025843 - | AATTAAGTTT-AAAGCGTTTTGGTTCGTCACAAGGGCAATTCTGCATT--TTTAAACTTGTATTGCACTTGAGACGGCCGAATTA--CTTTAAAAAGAA-----TC |
| droYak2 | chr2R:12724685-12724781 + | AATTGCGTTT-GAAGCGCTTCGGTTCGTCCCAGGAGCAATTCTGCACTA-TTAAAGCTAGTATTGCACTTGAGACGGCCGGATTA--CTTTTAAACGAA-----TC |
| droEre2 | scaffold_4845:10618147-10618242 - | AATTAAGTTT-GAAGCGCTTCGGTTCGTACCAGGAGCAATTCTGCATT--TTTAAGCTAGTATTGCACTTGAGACGGCCGGATTA--CTTTTAAACAAA-----AC |
| droAna3 | scaffold_13266:4132676-4132765 + | TCTACA--------ATTTTCCGGTGCGTCTCTAGTGCAACTCTTGGTTT-TTTATACAAGTATTGCACTTGAGACGGCCGAAATA--TAGTGGATCTAA-----GC |
| dp4 | chr3:8679756-8679828 + | G---------------------------------GGCAATACTGTGTTGTATTTCACCAGTATTGCACACCCACTGGCCTGAAAGTGCCTACTGCTGGGTTCAACA |
| droPer1 | gnl|ti|732455623:23-94 + | G---------------------------------GGCAATACTGTGTTGTATTTGAACAGTATTGCACACCCACTGGCCTGAAAGTGCCTACTGCTGGGTTCA--A |
| droWil1 | scaffold_180700:3453141-3453235 + | CATAAACTCGAAAATTTTTTAGGTTCGTTTAGTCTGCAAATT-GTAAT--ATACGAAAATAATTGCACACTTCACGGCCGGAAAA--GTTCAAGTTAAA-----CA |
| droGri2 | scaffold_15245:17564268-17564358 + | AAGCAAATCT-TCAGTTTTTCGGCTGTGGTTAGTGTCAATTCTTTTT---ATTTGACAAGCATTGCACTCTTCACGGCCGGATAA--A----TGAGGGT-----CT |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-19.9, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.1, p-value=0.009901 | dG=-19.1, p-value=0.009901 | dG=-19.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
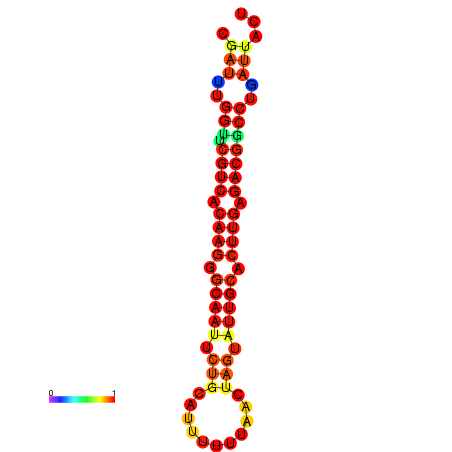
| dG=-20.4, p-value=0.009901 | dG=-20.4, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-19.7, p-value=0.009901 |
|---|---|---|---|---|
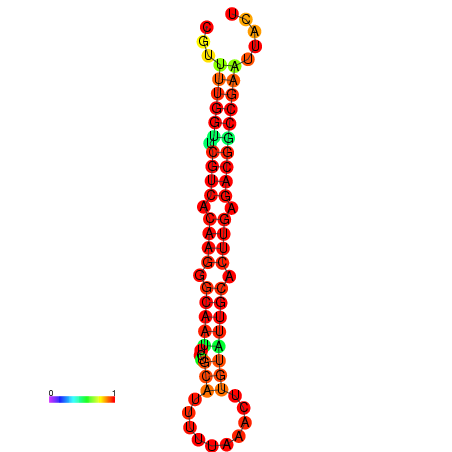 |
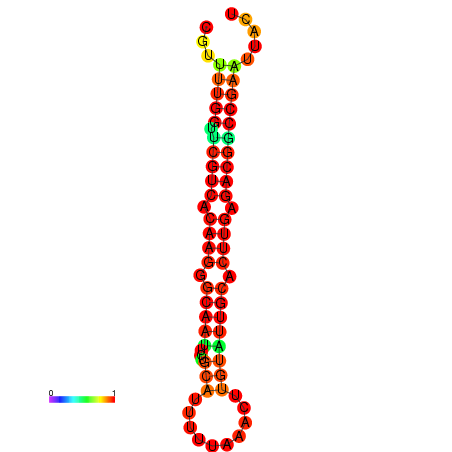 |
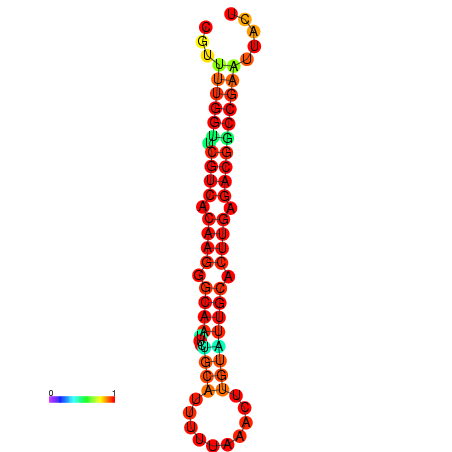 |
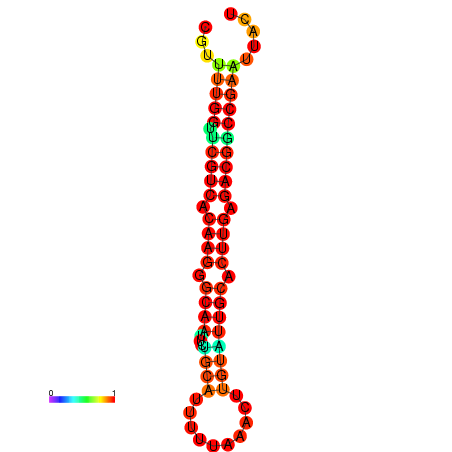 |
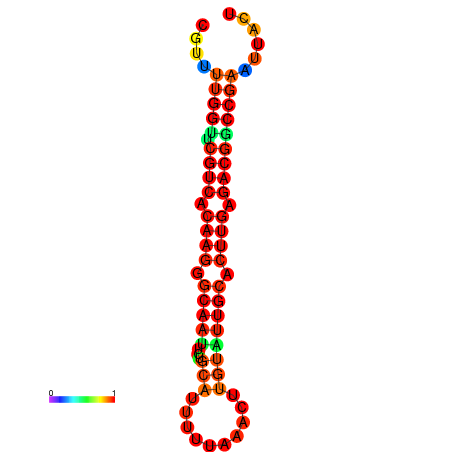 |
| dG=-20.4, p-value=0.009901 | dG=-20.4, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-19.7, p-value=0.009901 |
|---|---|---|---|---|
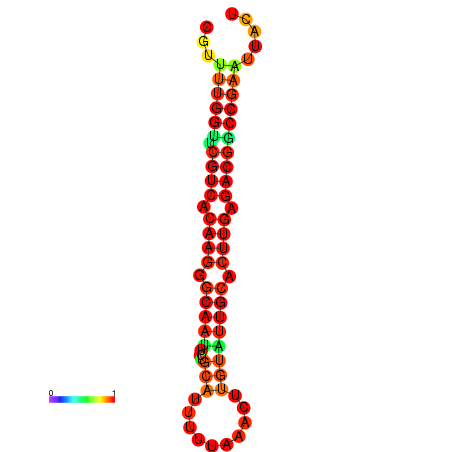 |
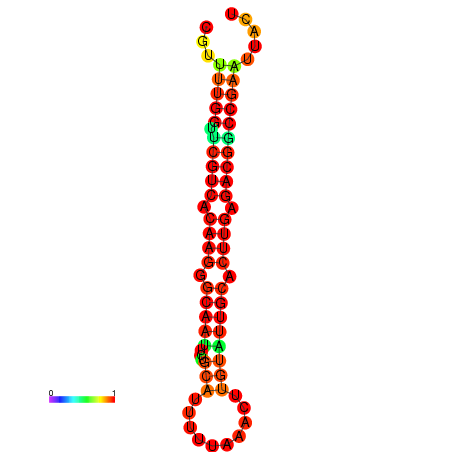 |
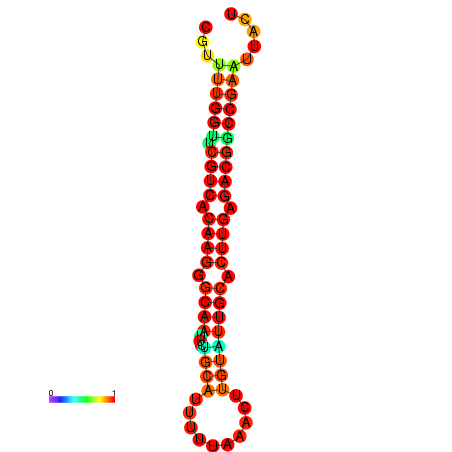 |
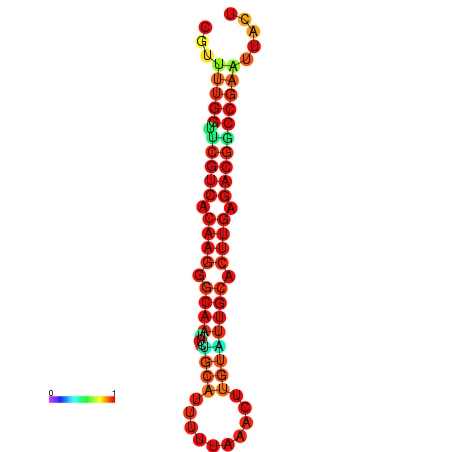 |
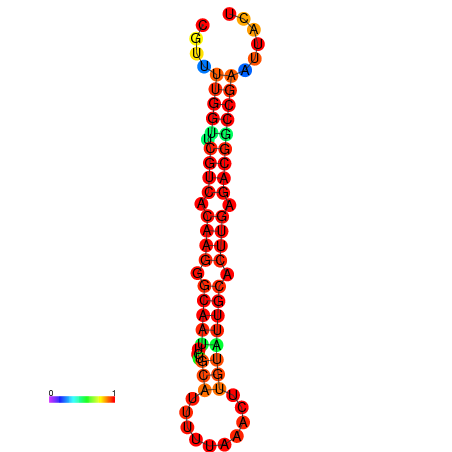 |
| dG=-24.5, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-23.7, p-value=0.009901 | dG=-23.7, p-value=0.009901 |
|---|---|---|---|
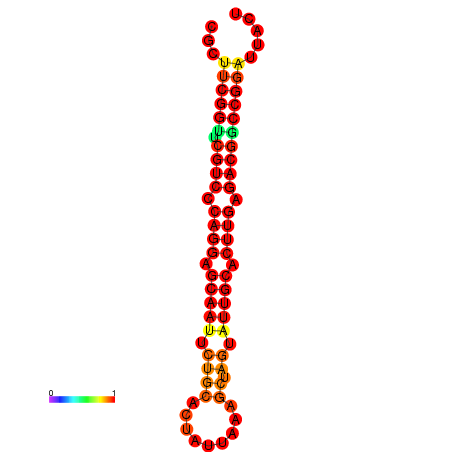 |
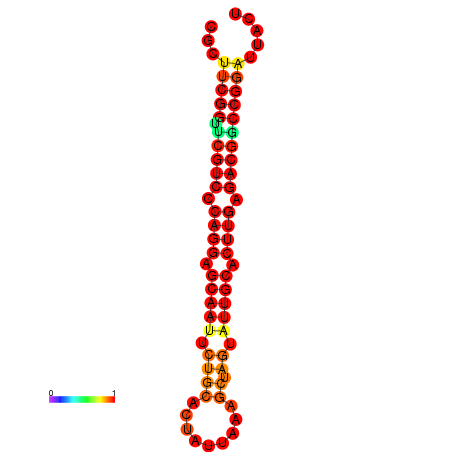 |
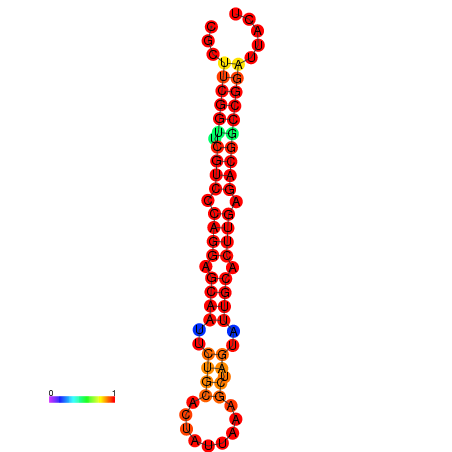 |
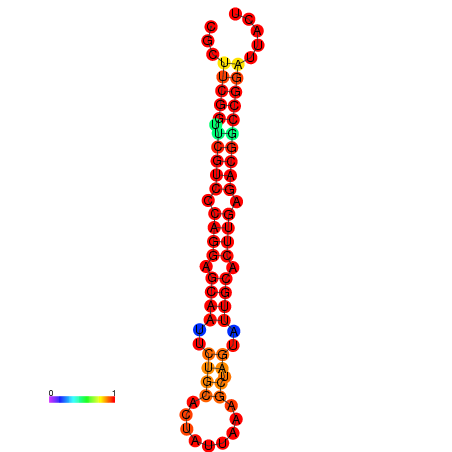 |
| dG=-22.4, p-value=0.009901 | dG=-22.4, p-value=0.009901 | dG=-21.6, p-value=0.009901 | dG=-21.6, p-value=0.009901 |
|---|---|---|---|
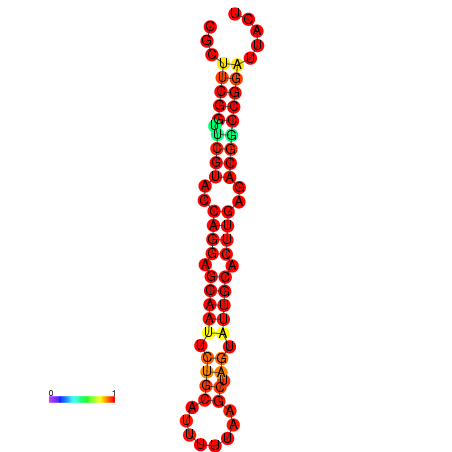
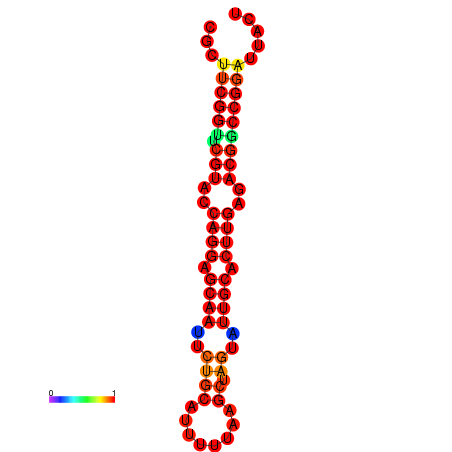
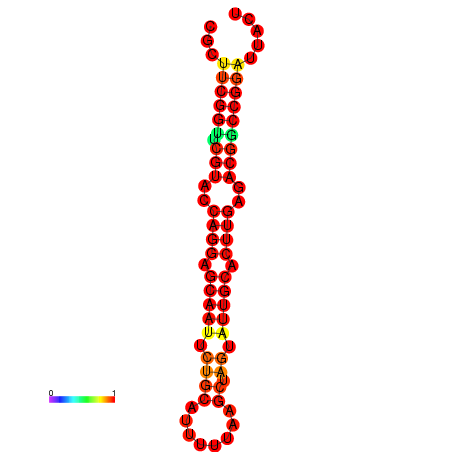 |
 |
 |
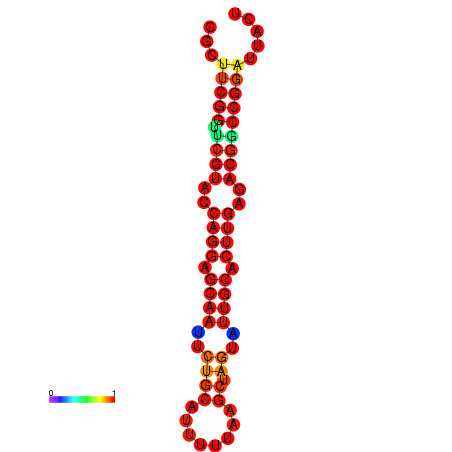 |
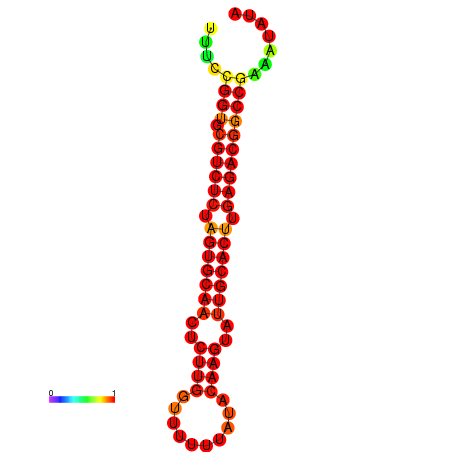
| dG=-30.2, p-value=0.009901 | dG=-29.2, p-value=0.009901 | dG=-29.2, p-value=0.009901 |
|---|---|---|
 |
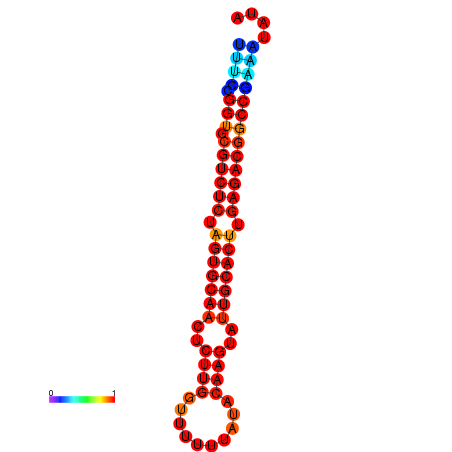 |
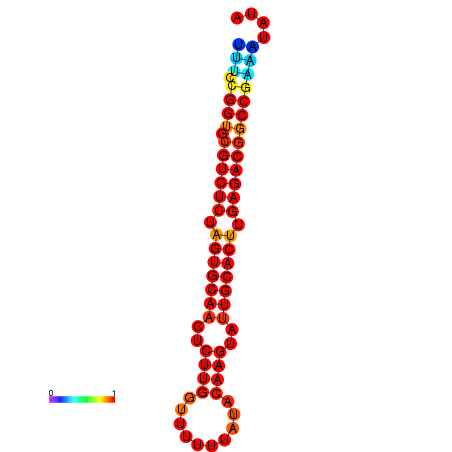 |
| dG=-16.9, p-value=0.009901 | dG=-16.9, p-value=0.009901 | dG=-16.6, p-value=0.009901 | dG=-15.9, p-value=0.009901 | dG=-15.9, p-value=0.009901 | dG=-13.4, p-value=0.009901 |
|---|---|---|---|---|---|
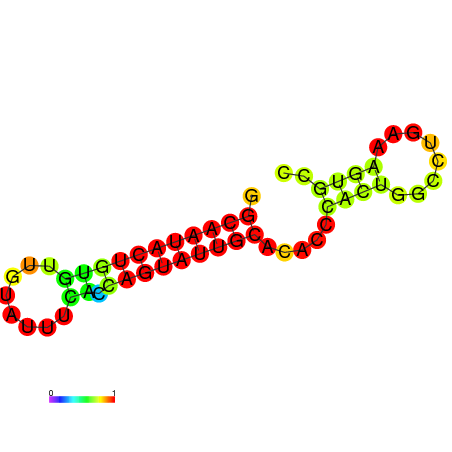 |
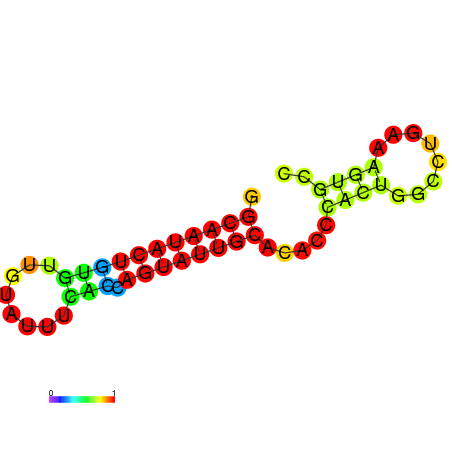 |
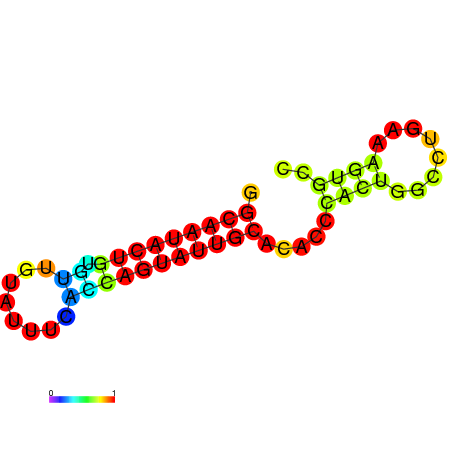 |
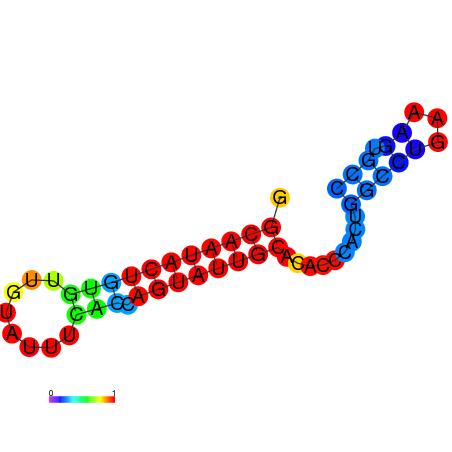 |
 |
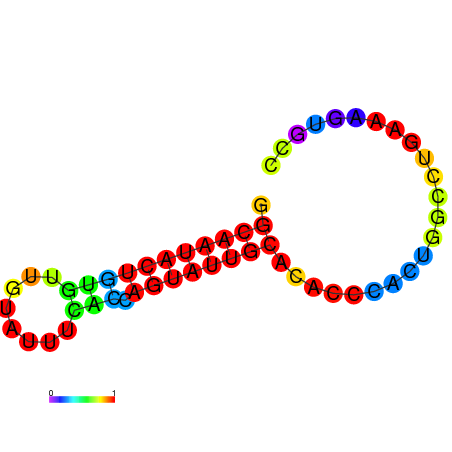 |
| dG=-17.4, p-value=0.009901 | dG=-16.4, p-value=0.009901 | dG=-13.9, p-value=0.009901 |
|---|---|---|
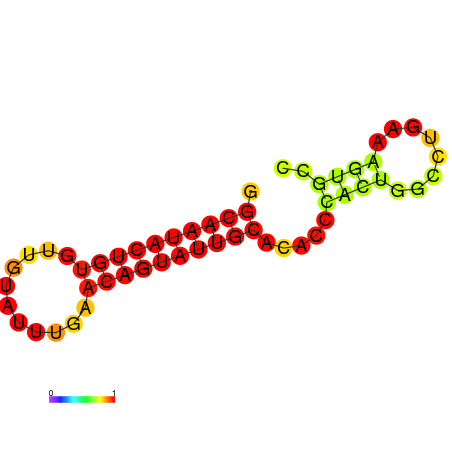 |
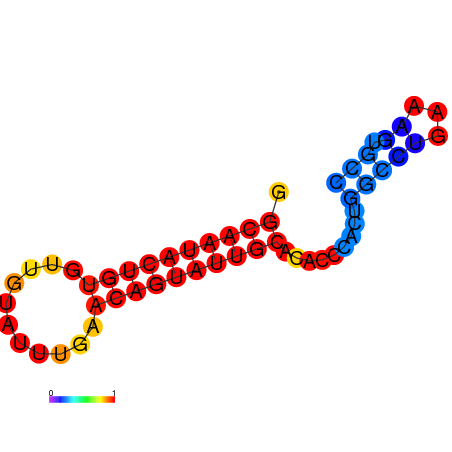 |
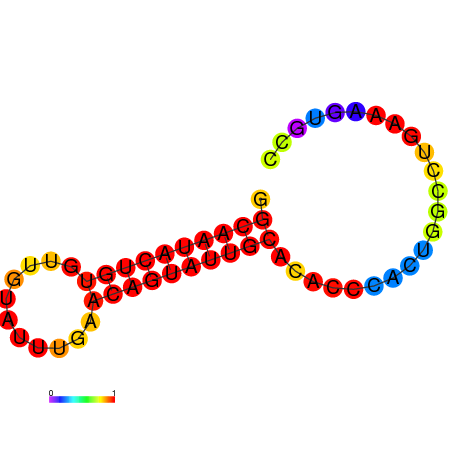 |
| dG=-14.6, p-value=0.009901 | dG=-14.6, p-value=0.009901 | dG=-14.6, p-value=0.009901 | dG=-14.6, p-value=0.009901 | dG=-14.6, p-value=0.009901 |
|---|---|---|---|---|
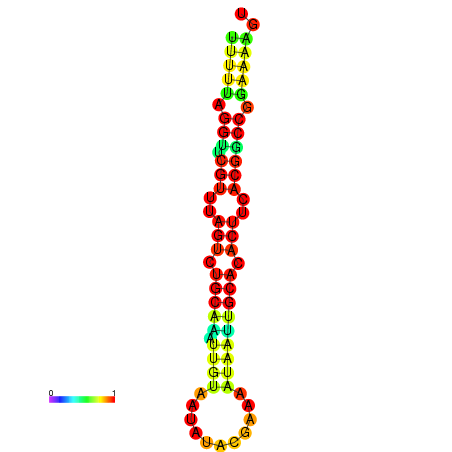 |
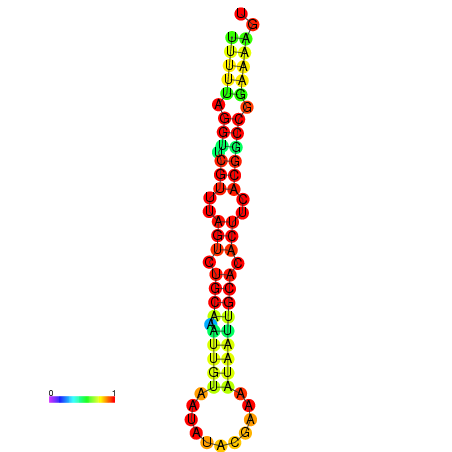 |
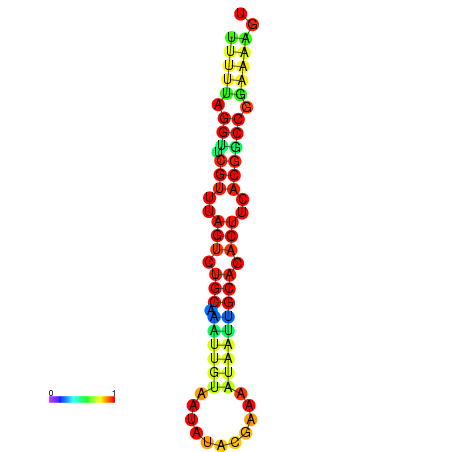 |
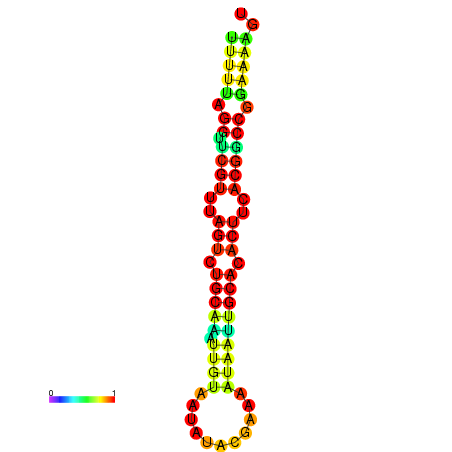 |
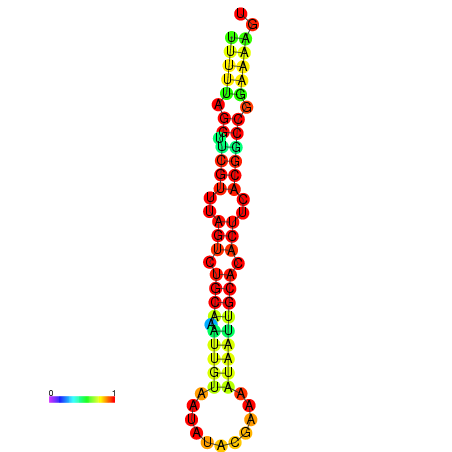 |
| dG=0.0, p-value=1.0 |
|---|
 |
| dG=0.0, p-value=1.0 |
|---|
 |
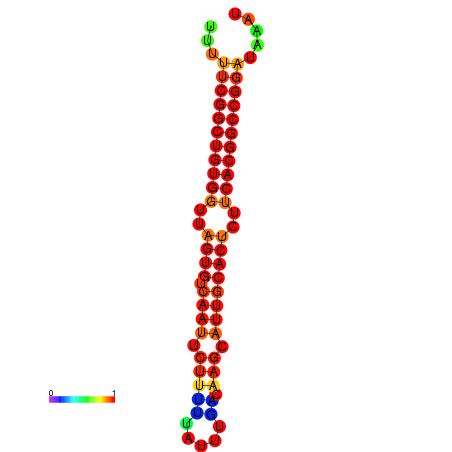
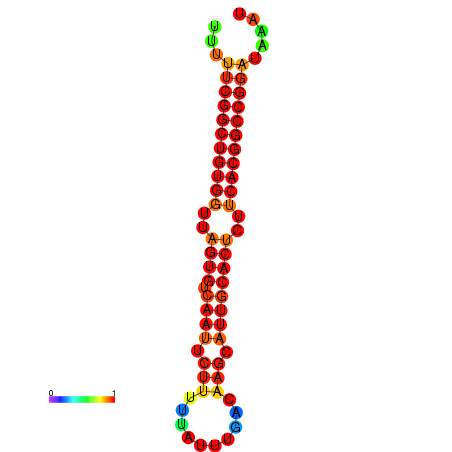
| dG=-24.4, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.5, p-value=0.009901 |
|---|---|---|---|---|
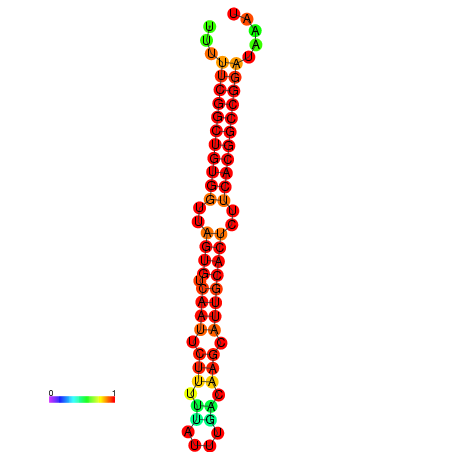 |
 |
 |
 |
 |
Generated: 03/07/2013 at 05:16 PM