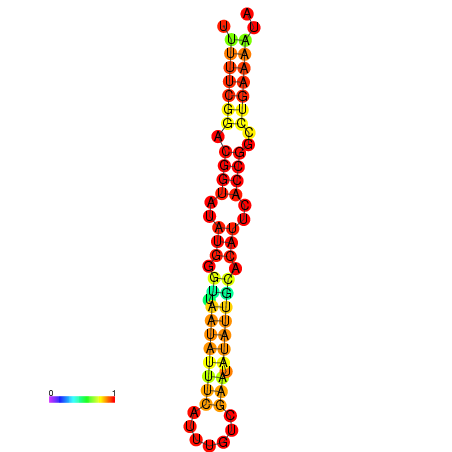
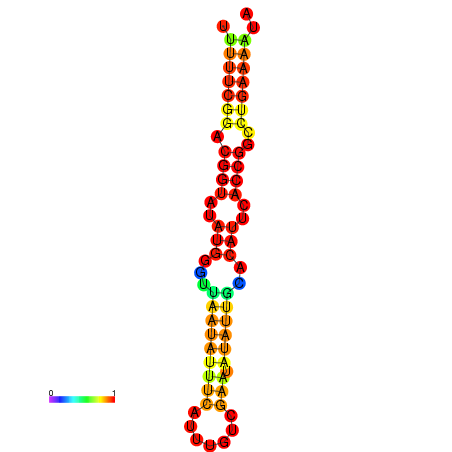
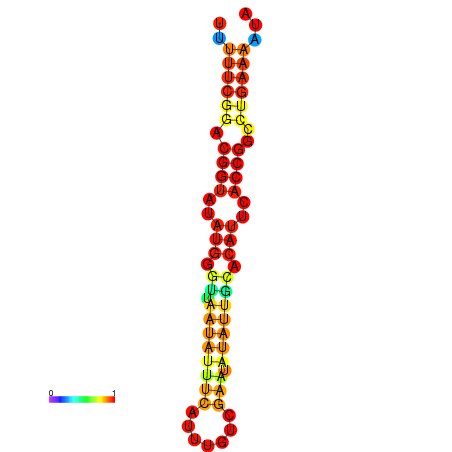
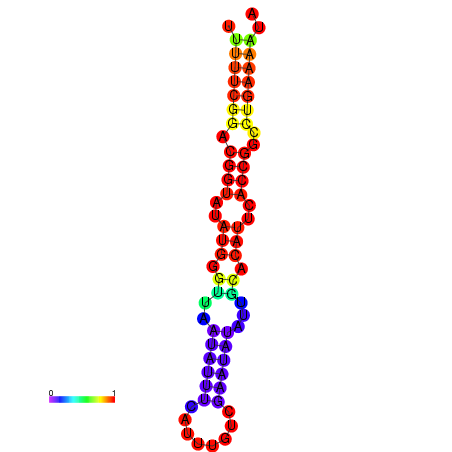
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16471367-16471480 - | TGAAAA-----------------A-TCG---AAATTTCTAGATCATTTTTCGGA--CGGTATATGGGTTAATATTTCAT------------TTGTCGAATATATTGCACATTCACCGGCCTGAAAATATCA-A--GAATT-------AACATC-----------------------------------GAA---AAG |
| droSim1 | chr2R:15132667-15132782 - | TGAAAA-----------------A-TCG---AAATTTCTAGATAATTTTTCGGA--CGGTATATGAGTCAATATTTTCT----------TTTTTTCGAATATATTGCACATTCACCGGCCTGAAAATATCA-A--GAATT-------AGCATC-----------------------------------GAA---AAA |
| droSec1 | super_1:14025591-14025696 - | GAAA------------------------------TTTCTAGATAATTTTTCGGA--CGGTATATGAGTCAATGTTTTCT------------TTTTCGAATATATTGCACATTCACCGGCCTGAAAATATCA-A--GAATT-------AGCATC-----------------------------------GAA---AAA |
| droYak2 | chr2R:12724807-12724923 + | TTTAAA-----------------AA-TATCTCACTTTCTAGATAGTTTTGCGGA--CGGTATCTGCGTCAATATTTCCT------------TGTTAGAATATATTGCACATTCACCGGCCTGAAAATATCA-A--GGACT-------TGCATC-----------------------------------GAA---AAG |
| droEre2 | scaffold_4845:10616532-10616644 - | TTGAAA-----------------C-CCC---TA-ACCCTAGATAGTTTTTCGGA--CGGTATCTGCGTCAATATTTCCT------------TGTTAGAATATATTGCACATTCACCGGCCTGAAAATATCA-A--GGACT-------TACATC-----------------------------------AAA---AAG |
| droAna3 | scaffold_13266:4132915-4133034 + | AGAGAA-----------------A-TAT---TTGTTGCCAGATTATCTTCAGGA--CGGTATTTGTGTCAATATGGATA-------------TTTCAAACATATTGCACTTATACCGGCCTGATGATCTCC-G--GCGCTCGTCATCTGTGAC-----------------------------------TAA---AAG |
| dp4 | chr3:8679876-8679985 + | CATCGA-----------------GC-TGCTTTGATTTGTA-----------GGC--CGTGGTTCTTGCAAATACGGATT---------------CATAACGTATTGCACTAGCCCCGGTCCAAAAAACAAT-A--GCAA-CGCC---GGCAAC-----------------------------------AGCAAAGTA |
| droPer1 | gnl|ti|732560583:591-697 + | CATCGA-----------------GC-TGCTTTGATTTGTA-----------GGC--CGTGGTTCTTGCAAATACGAATT---------------CATAACGTATTGCACTAGCCCCGGTCCAAAAAACAAT-A--GCAA-CGCC---GGCAAC-----------------------------------AGC---AAA |
| droWil1 | scaffold_180700:3453290-3453392 + | TAAT-------------------------------TTATA-----------GGCTTCGGGGT-TGTTGCAAAATGGTTTATTCT-----AATAACTCCCCATATTGCACCTTTTCCGGCCTCTAAATTAACTTTTG-----------AACATCCCG--------------------------------GGT---GGG |
| droVir3 | scaffold_12875:13668876-13669002 - | GAAAAA-----------------C-TAG-TGCGTCCGCATGACCATTTTAAGGA--CGGTATTTGTGACAATATGTGCAG-TTAAATCAATTTGTTTAGCATATTGCACATCTCCCGGCCTAAAAATGAAC-C--GTGCT-------CTCTTC-----------------------------------GAT---ATA |
| droMoj3 | scaffold_6496:9090405-9090568 - | AAACGAAAAATGTCAAATTTTCAGC-TG---------------CGTTTGCAGGA--CGGTATTTGTGACAATATGAATTATTGAAATCAATTTATTCAGCATATTGCACATCTCCCGGCCTATAAAACAGT-T--GTGA-GCCC---GTCGAT---TTAAATGATTTGTGTATTTAAAATAAATTTTCACC-----A |
| droGri2 | scaffold_15245:17564385-17564474 + | AAAT-------------------------------TTGCAGTTCGTGTTTAG------T-----------GTTTTTTCC------------CGCTTAAGCATCTTGCAAATTATATGGCTTGCAGTTATCT-G--CATTA-------ACCATT-----------------------------------TTC---AGG |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16471367-16471480 - | TGAAAA-----------------A-TCG---AAATTTCTAGATCATTTTTCGGA--CGGTATATGGGTTAATATTTCATT------------TGTCGAATATATTGCACATTCACCGGCCTGAAAAT-----ATCAAGAATT-------A-----------------------------------ACATCGAAAAG |
| droSim1 | chr2R:15132667-15132782 - | TGAAAA-----------------A-TCG---AAATTTCTAGATAATTTTTCGGA--CGGTATATGAGTCAATATTTTCTT----------TTTTTCGAATATATTGCACATTCACCGGCCTGAAAAT-----ATCAAGAATT-------A-----------------------------------GCATCGAAAAA |
| droSec1 | super_1:14025591-14025696 - | GAA------------------------------ATTTCTAGATAATTTTTCGGA--CGGTATATGAGTCAATGTTTTCTT------------TTTCGAATATATTGCACATTCACCGGCCTGAAAAT-----ATCAAGAATT-------A-----------------------------------GCATCGAAAAA |
| droYak2 | chr2R:12724807-12724923 + | TTTAAA-----------------AA-TATCTCACTTTCTAGATAGTTTTGCGGA--CGGTATCTGCGTCAATATTTCCTT------------GTTAGAATATATTGCACATTCACCGGCCTGAAAAT-----ATCAAGGACT-------T-----------------------------------GCATCGAAAAG |
| droEre2 | scaffold_4845:10616532-10616644 - | TTGAAA-----------------C-CCC---TA-ACCCTAGATAGTTTTTCGGA--CGGTATCTGCGTCAATATTTCCTT------------GTTAGAATATATTGCACATTCACCGGCCTGAAAAT-----ATCAAGGACT-------T-----------------------------------ACATCAAAAAG |
| droAna3 | scaffold_13266:4132915-4133034 + | AGAGAA-----------------A-TAT---TTGTTGCCAGATTATCTTCAGGA--CGGTATTTGTGTCAATATGGATAT-------------TTCAAACATATTGCACTTATACCGGCCTGATGAT-----CTCCGGCGCTCGTCATCT-----------------------------------GTGACTAAAAG |
| dp4 | chr3:8679876-8679985 + | CATCGA-----------------GC-TGCTTTGATTTG-----------TAGGC--CGTGGTTCTTGCAAATACGGATT---------------CATAACGTATTGCACTAGCCCCGGTCCAAAAAACAAT-AGCAACGC-C-------GGCA--------------------------------ACAGCAAAGTA |
| droPer1 | gnl|ti|732560583:591-697 + | CATCGA-----------------GC-TGCTTTGATTTG-----------TAGGC--CGTGGTTCTTGCAAATACGAATT---------------CATAACGTATTGCACTAGCCCCGGTCCAAAAAACAAT-AGCAAC-GCC-------G-----------------------------------GCAACAGCAAA |
| droWil1 | scaffold_180700:3453290-3453392 + | TAATTT-------------------------------A-----------TAGGCTTCGGGGT-TGTTGCAAAATGGTTTATTCTA-----ATAACTCCCCATATTGCACCTTTTCCGGCCTCTAAATTAACTTTTGAACA---------------------------------------------TCCCGGGTGGG |
| droVir3 | scaffold_12875:13668876-13669002 - | GAAAAA-----------------C-TAG-TGCGTCCGCATGACCATTTTAAGGA--CGGTATTTGTGACAATATGTGCAG-TTAAATCAATTTGTTTAGCATATTGCACATCTCCCGGCCTAAAAAT-----GAACCGTGCT-------C-----------------------------------TCTTCGATATA |
| droMoj3 | scaffold_6496:9090405-9090568 - | AAACGAAAAATGTCAAATTTTCAGC-TG---------------CGTTTGCAGGA--CGGTATTTGTGACAATATGAATTATTGAAATCAATTTATTCAGCATATTGCACATCTCCCGGCCTATAAAACAGT-TGTGAGCC-C-------GTCGATTTAAATGATTTGTGTATTTAAAATAAATTTTCACC-----A |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-16.2, p-value=0.009901 | dG=-15.9, p-value=0.009901 | dG=-15.5, p-value=0.009901 | dG=-15.3, p-value=0.009901 | dG=-15.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-18.3, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.3, p-value=0.009901 | dG=-17.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
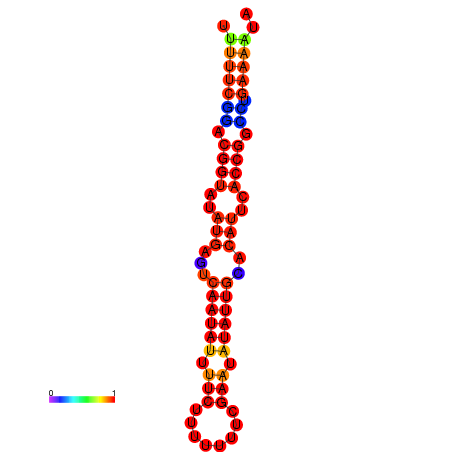 |
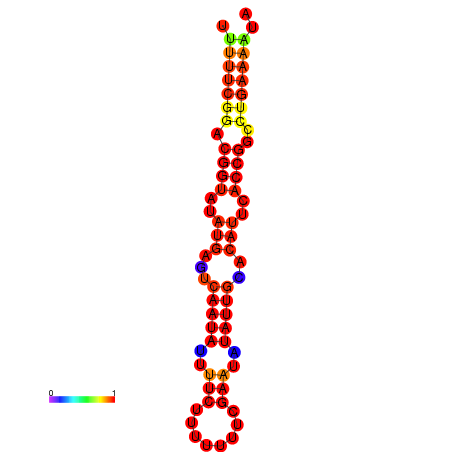 |
| dG=-18.4, p-value=0.009901 | dG=-17.7, p-value=0.009901 | dG=-17.7, p-value=0.009901 | dG=-17.4, p-value=0.009901 |
|---|---|---|---|
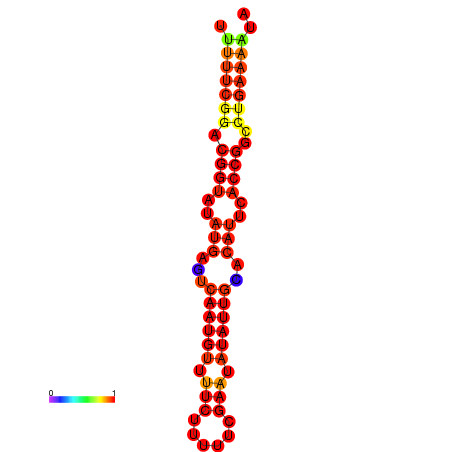 |
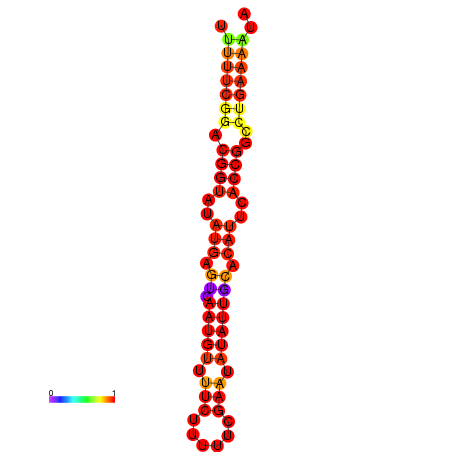 |
 |
 |
| dG=-11.5, p-value=0.009901 | dG=-11.2, p-value=0.009901 | dG=-11.2, p-value=0.009901 | dG=-11.1, p-value=0.009901 | dG=-11.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
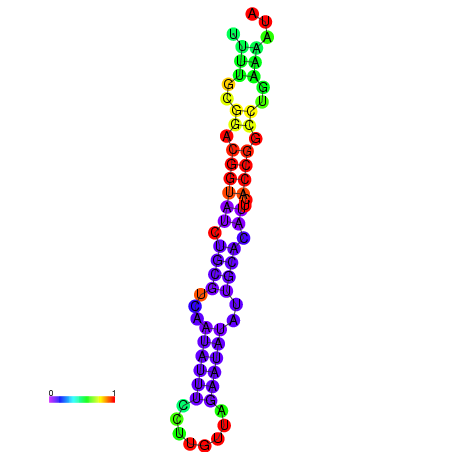 |
 |
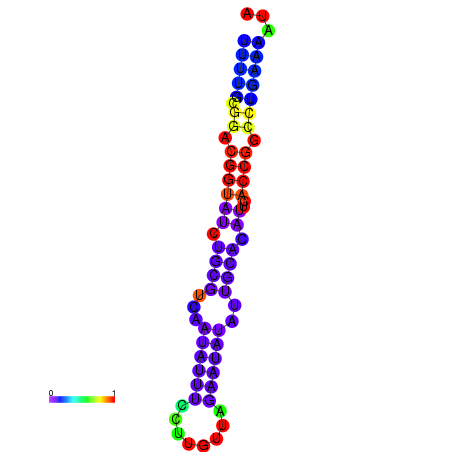 |
| dG=-14.4, p-value=0.009901 | dG=-14.1, p-value=0.009901 | dG=-14.0, p-value=0.009901 | dG=-13.9, p-value=0.009901 | dG=-13.7, p-value=0.009901 |
|---|---|---|---|---|
 |
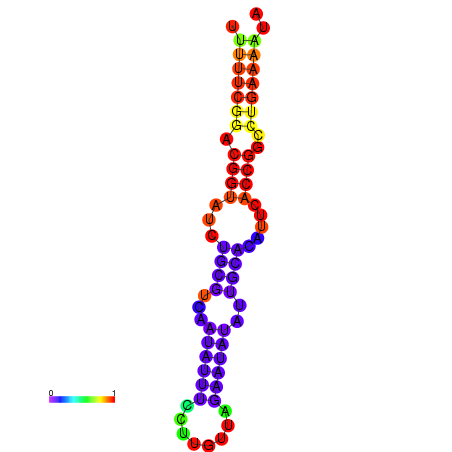 |
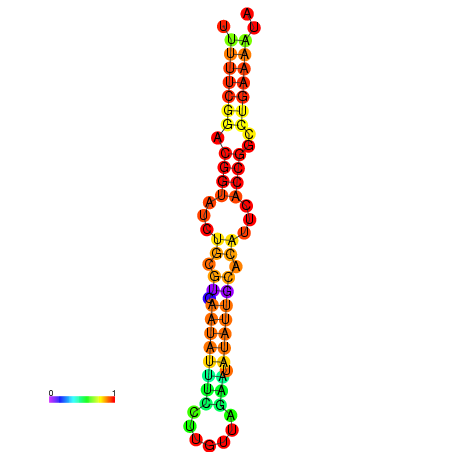 |
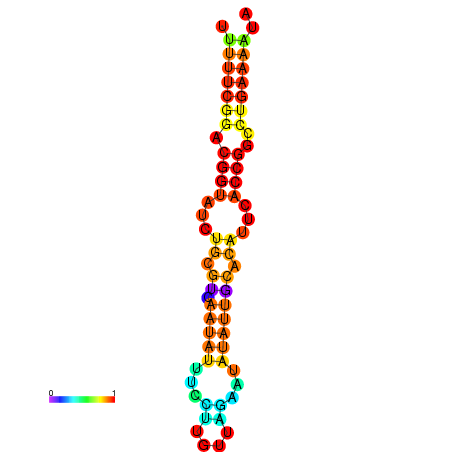 |
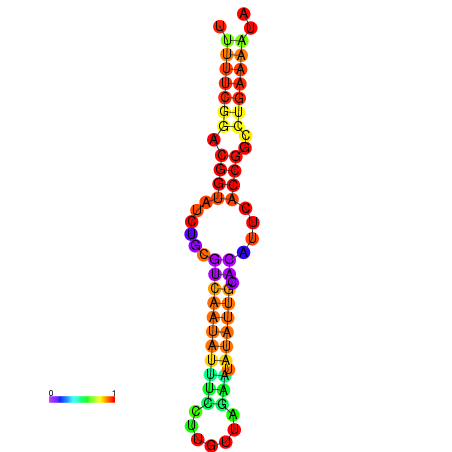 |
| dG=-29.0, p-value=0.009901 |
|---|
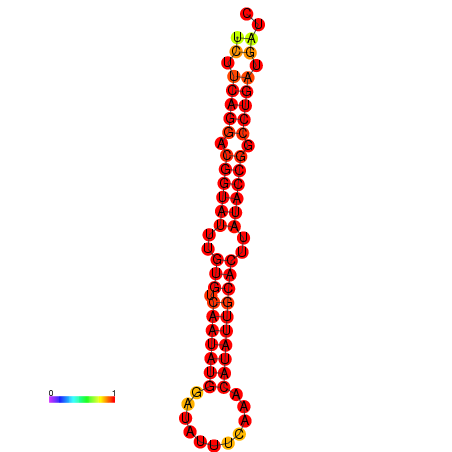 |
| dG=-21.4, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.0, p-value=0.009901 | dG=-21.0, p-value=0.009901 |
|---|---|---|---|---|
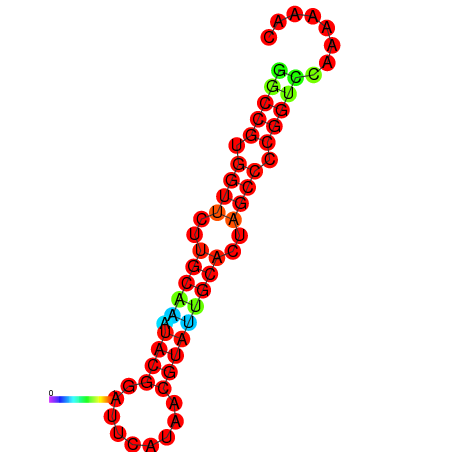 |
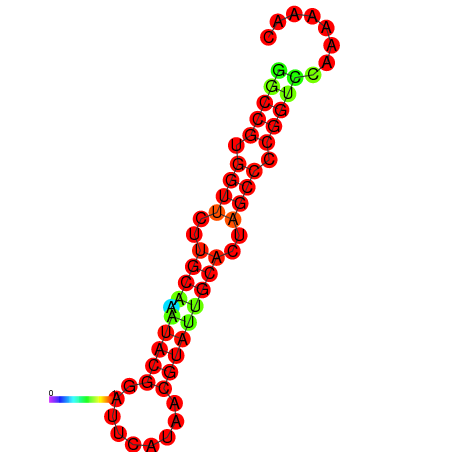 |
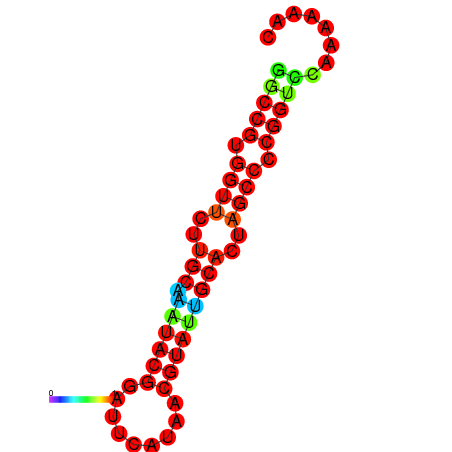 |
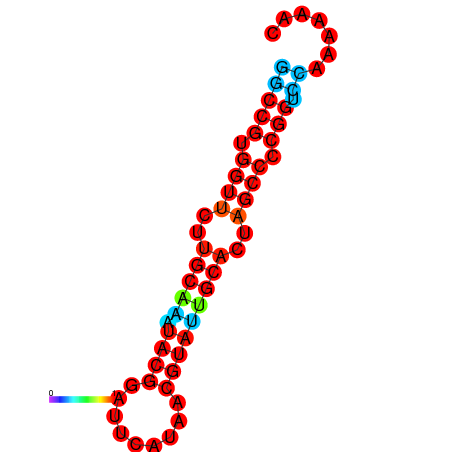 |
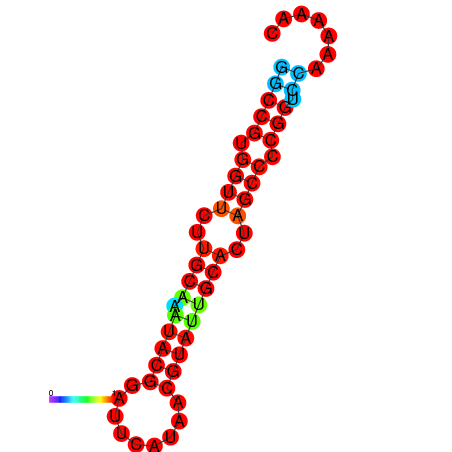 |
| dG=-20.0, p-value=0.009901 | dG=-20.0, p-value=0.009901 | dG=-20.0, p-value=0.009901 | dG=-19.6, p-value=0.009901 | dG=-19.6, p-value=0.009901 |
|---|---|---|---|---|
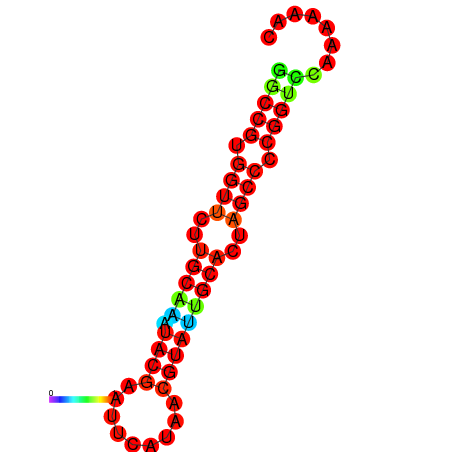 |
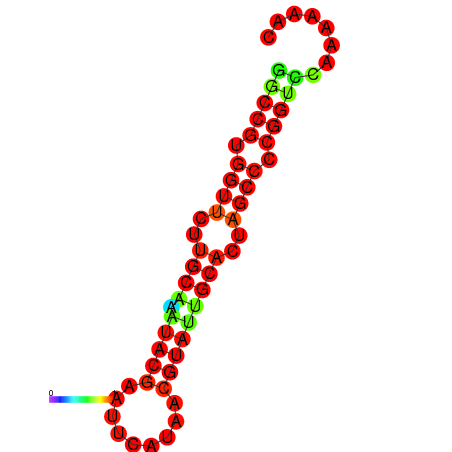 |
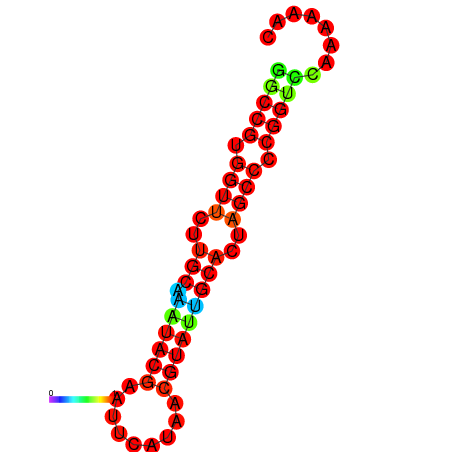 |
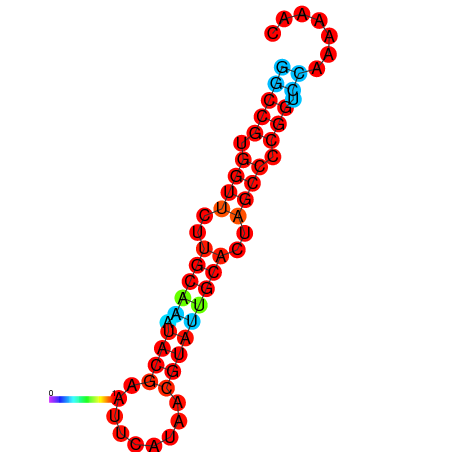 |
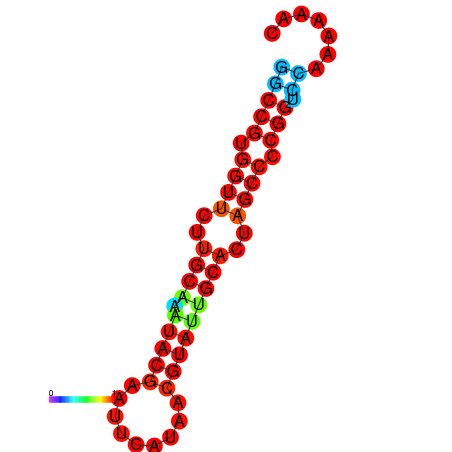 |
| dG=-16.1, p-value=0.009901 | dG=-15.6, p-value=0.009901 | dG=-15.3, p-value=0.009901 |
|---|---|---|
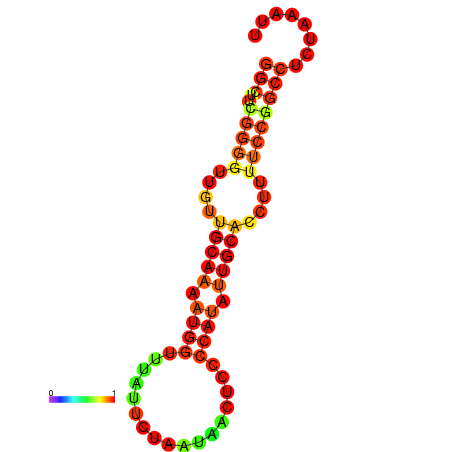 |
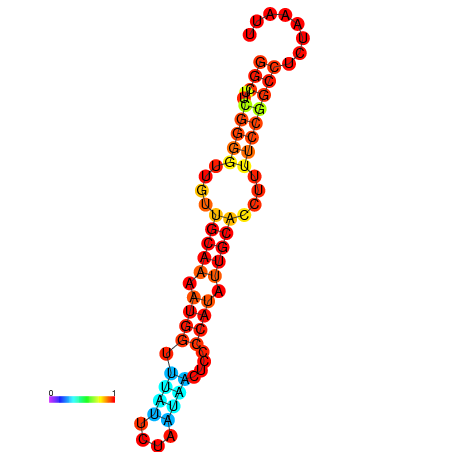 |
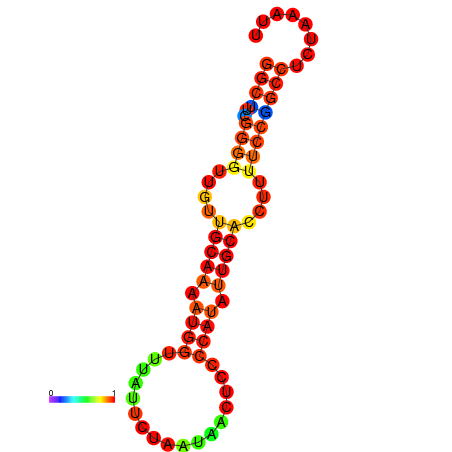 |
| dG=-21.2, p-value=0.009901 | dG=-21.2, p-value=0.009901 | dG=-21.2, p-value=0.009901 | dG=-21.1, p-value=0.009901 | dG=-20.9, p-value=0.009901 |
|---|---|---|---|---|
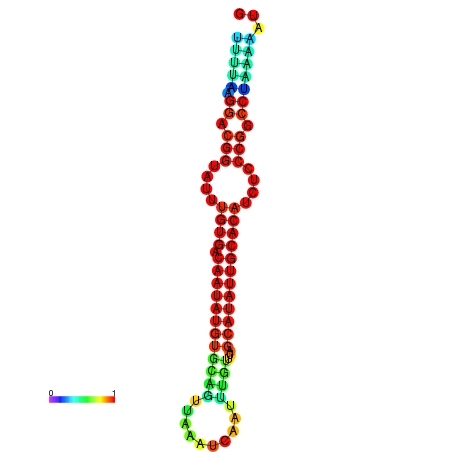 |
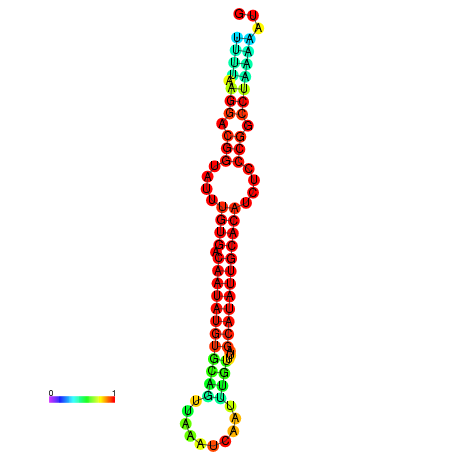 |
 |
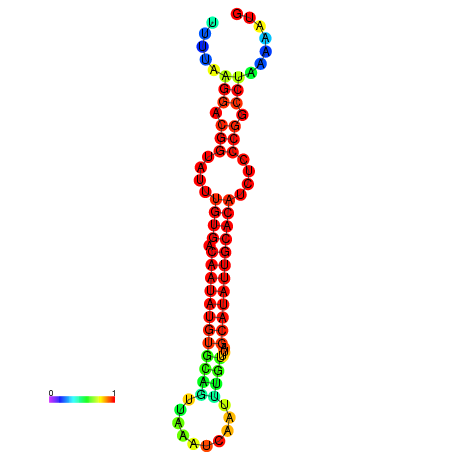 |
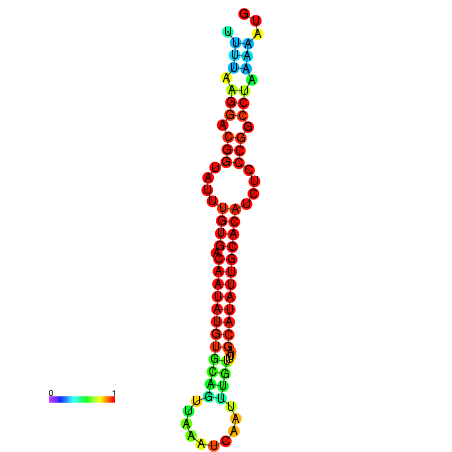 |
| dG=-20.0, p-value=0.009901 | dG=-19.5, p-value=0.009901 | dG=-19.5, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-7.8, p-value=0.009901 | dG=-7.8, p-value=0.009901 | dG=-7.4, p-value=0.009901 | dG=-7.4, p-value=0.009901 | dG=-7.2, p-value=0.009901 | dG=-4.7, p-value=0.009901 |
|---|---|---|---|---|---|
 |
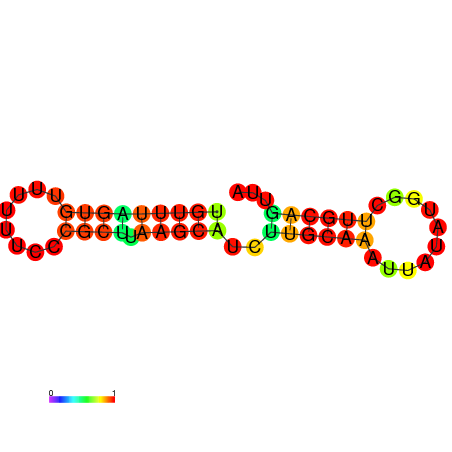 |
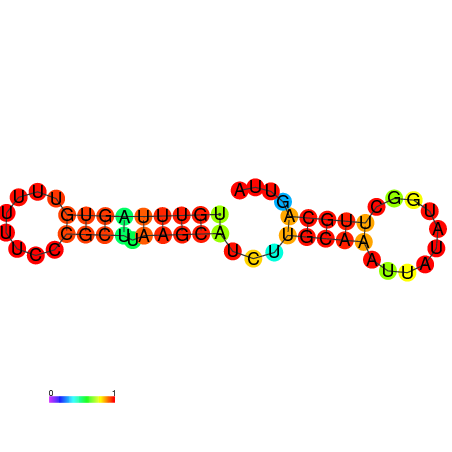 |
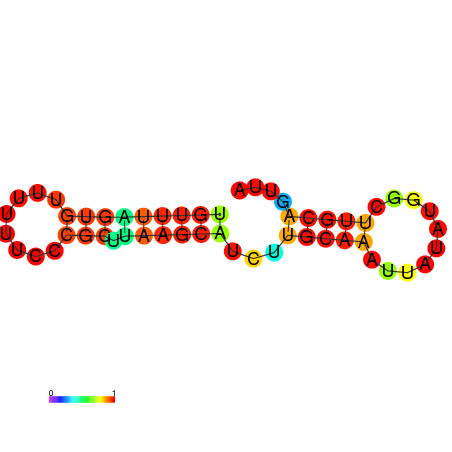 |
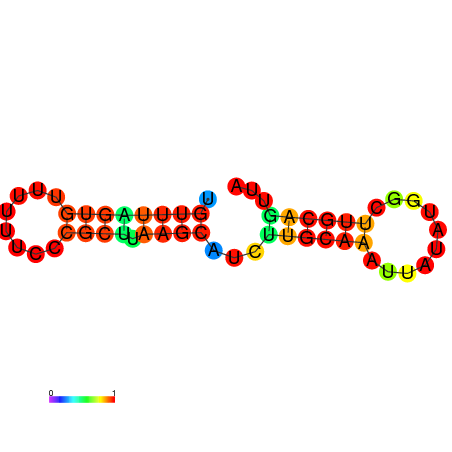 |
 |
Generated: 03/07/2013 at 05:16 PM