
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16471242-16471360 - | AAAAGTGTTT------GC----G-----------------------AAAACATAAACATTTGCAGGGC-GGGTCGTGTGTCAGTGTATTTAT-------------ATCT-TAGC----------TATATTGCACACTTCCCGGCCTTTAAATGTCCA---ATGTTTCAACATT-------------TAAATCT----- |
| droSim1 | chr2R:15132542-15132660 - | AAAAGTGCTT------GC----A-----------------------AAAACATAAACATTTGCAGGAC-GGGTCGTGTGTCAGTGTATTTAC-------------TTCT-TAGC----------TATATTGCACACTTCCCGGCCTTTAAATGTCCA---ATGTTTCAACTTT-------------AATATCT----- |
| droSec1 | super_1:14025465-14025583 - | AAAAGTGCTT------GC----A-----------------------AAAACATAAACATTTGCAGGAC-GGGTCGTGTGTCAGTGTATTTAC-------------TTCT-TAAC----------TATATTGCACACTTCCCGGCCTTTAAATGTCCA---ATGTTTCAACTTT-------------AATATCT----- |
| droYak2 | chr2R:12724925-12725037 + | CTT-------------GC----A-----------------------AATACACAAACATTTGCAGGAC-GGGTCGTGTGGCAGTGTATTGAT-------------TTCT-TAGC----------TATATTGCACACTTCCCGCCCTTTAAATGTCCA---GTGTTTCGACTTG-------------GATATCT----- |
| droEre2 | scaffold_4845:10616419-10616531 - | CTT-------------GC----A-----------------------AATACACAAATATTTGCAGGAC-GGGTCGTGTGTCAATGTATTGAT-------------CTCT-TAGC----------TATATTGCACACTTCCCGCCCTTTAAATGTCCA---GTGTTTCAACTTG-------------AATATCT----- |
| droAna3 | scaffold_13266:4133041-4133158 + | AATAGC----------------------------------------AAAACATAAACACTTGCAGGAC-GGGGAGCGTGACAATATATATAT-------------TTTA-TAAA--------CATATATTGCACACCTCCCGGCCTATAAGTGTAAA----TGTTCCTACAGG-------------TAAAACTTCTAA |
| dp4 | chr3:8679992-8680133 + | ATTATAGTGT------GCGGAGTATGTGCTGTGTAT--T-------TGTACACCAACATTTGTGCGTCTAAGATCTGT-GCAGTATATACAT-------ACATAG-AA-TACAA----------AATATTGCAGACTTCTCAGACTACAAATGTGAA---CTGTCTGGTCTTC-------------AAAAAAT----- |
| droPer1 | super_4:2566796-2566912 + | CTGTGCATTT------G-------------------------------TACACCAACATTTGTTCGTCTAAGTTCTGTG-CAGTATATACAT-------ACATAG-TT-TAAAA----------AATATTGCACACTTCTCAGACTACAAATGTGAA---CTGTCTGGTCTTC-------------AAAA-------- |
| droWil1 | scaffold_180700:3453399-3453550 + | GAAGGAGTCAATGACCAC----A-----------------------AATTTTCAAACATTCTTTGGAC-GGACTT-GT-CAAATATGTTTGG-------ATATAAAATT-CAACAATTGGACAACATATTGATCAAAACCGGGCCAAAAATTGTTGG---AAAATTCTTCAAAGTTGTTGGACTTGACAATTT----- |
| droVir3 | scaffold_12875:13668721-13668866 - | AAAACAATTT------GG----TAAATACTGCGCATAGCCTTGCGCCTTATGTAATCATTTTTAGGCC-GTGCTCTGTGTGAATATGTTTTT-------------AACTATAAT----------CATATTGCACATCTCCCGGCCTATAAGTGTAGATGCATAATTCATCAAA-------------TATAAAT----- |
| droMoj3 | scaffold_6496:9090283-9090389 - | TGT--------------------------------------------------ATCCATTTTTAGGCC-GTGCTATGTGACAATATGTATTT-------AAATTG-TA-TTTAT----------CATATTGCACATCTCCCGGCCTACAAGTGTAAATGCACAATTCAACT----------------ATGTAC----- |
| droGri2 | scaffold_15245:17564471-17564548 + | C--------------------------------------------------------------AGGAT-GGAAGCTGTGACAATATGTAGGTTTTAATTAATTTA-TA-TTTCA----------TATATTGCACACCTCCCGGCCTATAAATG--------------------------------------------- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16471242-16471360 - | AAAAGTGTTT------GC----G-----------------------AAAACATAAACATTTGCAGGGC-GGGTCGTGTGTCAGTGTATTTAT-------------ATCT-TAGC----------TATATTGCACACTTCCCGGCCTTTAAATGTCCA---ATGTTTCAACATT-------------TAAATCT----- |
| droSim1 | chr2R:15132542-15132660 - | AAAAGTGCTT------GC----A-----------------------AAAACATAAACATTTGCAGGAC-GGGTCGTGTGTCAGTGTATTTAC-------------TTCT-TAGC----------TATATTGCACACTTCCCGGCCTTTAAATGTCCA---ATGTTTCAACTTT-------------AATATCT----- |
| droSec1 | super_1:14025465-14025583 - | AAAAGTGCTT------GC----A-----------------------AAAACATAAACATTTGCAGGAC-GGGTCGTGTGTCAGTGTATTTAC-------------TTCT-TAAC----------TATATTGCACACTTCCCGGCCTTTAAATGTCCA---ATGTTTCAACTTT-------------AATATCT----- |
| droYak2 | chr2R:12724925-12725037 + | CTT-------------GC----A-----------------------AATACACAAACATTTGCAGGAC-GGGTCGTGTGGCAGTGTATTGAT-------------TTCT-TAGC----------TATATTGCACACTTCCCGCCCTTTAAATGTCCA---GTGTTTCGACTTG-------------GATATCT----- |
| droEre2 | scaffold_4845:10616419-10616531 - | CTT-------------GC----A-----------------------AATACACAAATATTTGCAGGAC-GGGTCGTGTGTCAATGTATTGAT-------------CTCT-TAGC----------TATATTGCACACTTCCCGCCCTTTAAATGTCCA---GTGTTTCAACTTG-------------AATATCT----- |
| droAna3 | scaffold_13266:4133041-4133158 + | AATAGC----------------------------------------AAAACATAAACACTTGCAGGAC-GGGGAGCGTGACAATATATATAT-------------TTTA-TAAA--------CATATATTGCACACCTCCCGGCCTATAAGTGTAAA----TGTTCCTACAGG-------------TAAAACTTCTAA |
| dp4 | chr3:8679992-8680133 + | ATTATAGTGT------GCGGAGTATGTGCTGTGTAT--T-------TGTACACCAACATTTGTGCGTCTAAGATCTGT-GCAGTATATACAT-------ACATAG-AA-TACAA----------AATATTGCAGACTTCTCAGACTACAAATGTGAA---CTGTCTGGTCTTC-------------AAAAAAT----- |
| droPer1 | super_4:2566796-2566912 + | CTGTGCATTT------G-------------------------------TACACCAACATTTGTTCGTCTAAGTTCTGTG-CAGTATATACAT-------ACATAG-TT-TAAAA----------AATATTGCACACTTCTCAGACTACAAATGTGAA---CTGTCTGGTCTTC-------------AAAA-------- |
| droWil1 | scaffold_180700:3453399-3453550 + | GAAGGAGTCAATGACCAC----A-----------------------AATTTTCAAACATTCTTTGGAC-GGACTT-GT-CAAATATGTTTGG-------ATATAAAATT-CAACAATTGGACAACATATTGATCAAAACCGGGCCAAAAATTGTTGG---AAAATTCTTCAAAGTTGTTGGACTTGACAATTT----- |
| droVir3 | scaffold_12875:13668721-13668866 - | AAAACAATTT------GG----TAAATACTGCGCATAGCCTTGCGCCTTATGTAATCATTTTTAGGCC-GTGCTCTGTGTGAATATGTTTTT-------------AACTATAAT----------CATATTGCACATCTCCCGGCCTATAAGTGTAGATGCATAATTCATCAAA-------------TATAAAT----- |
| droMoj3 | scaffold_6496:9090283-9090389 - | TGT--------------------------------------------------ATCCATTTTTAGGCC-GTGCTATGTGACAATATGTATTT-------AAATTG-TA-TTTAT----------CATATTGCACATCTCCCGGCCTACAAGTGTAAATGCACAATTCAACT----------------ATGTAC----- |
| droGri2 | scaffold_15245:17564471-17564548 + | C--------------------------------------------------------------AGGAT-GGAAGCTGTGACAATATGTAGGTTTTAATTAATTTA-TA-TTTCA----------TATATTGCACACCTCCCGGCCTATAAATG--------------------------------------------- |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-28.3, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-28.0, p-value=0.009901 |
|---|---|---|
 |
 |
 |
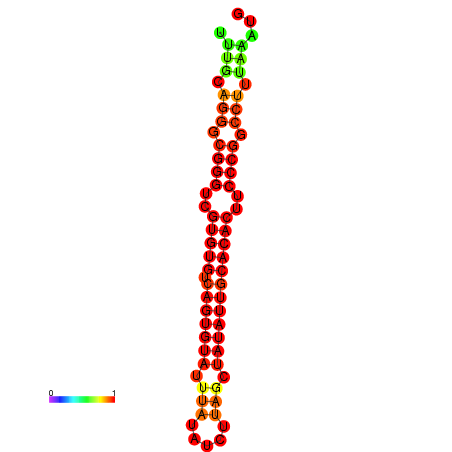
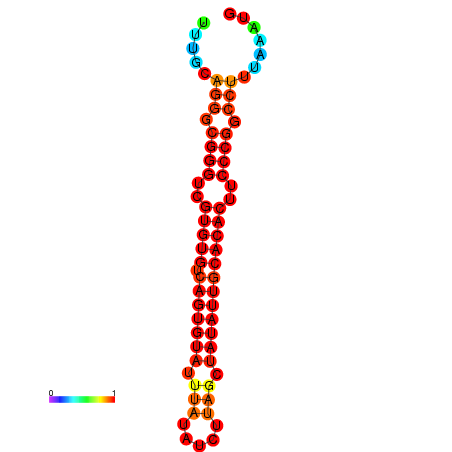
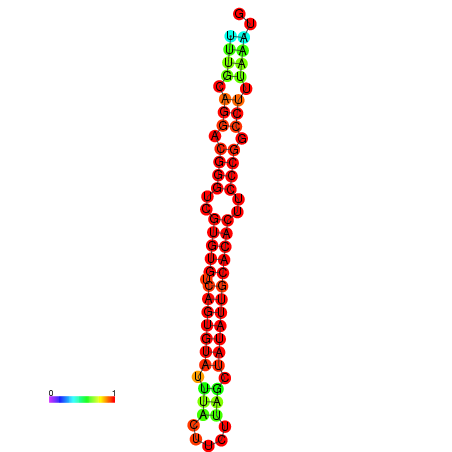
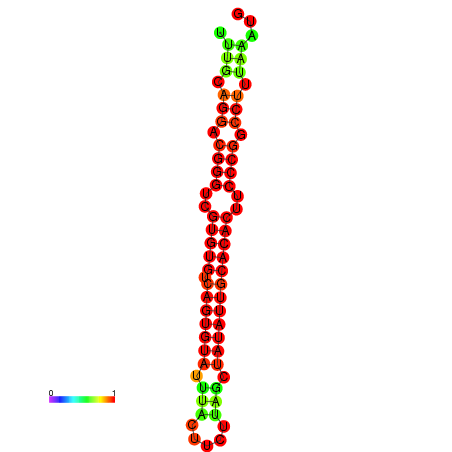
| dG=-24.5, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-23.6, p-value=0.009901 | dG=-23.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
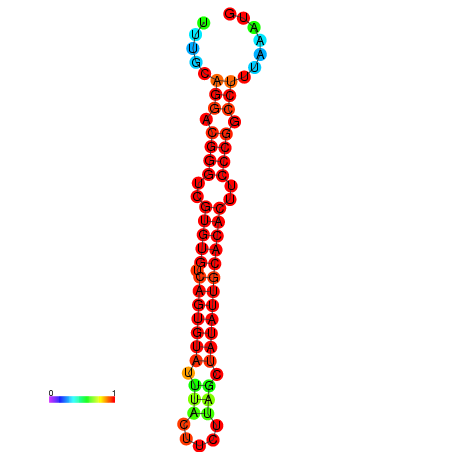 |
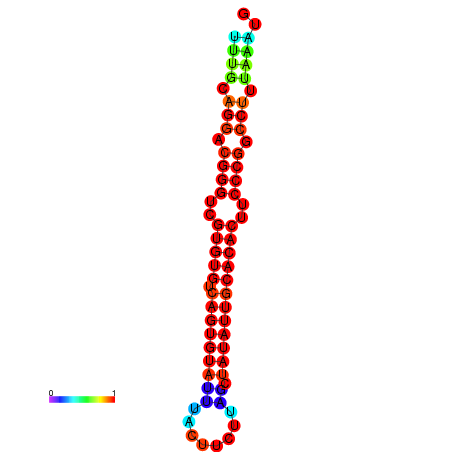 |
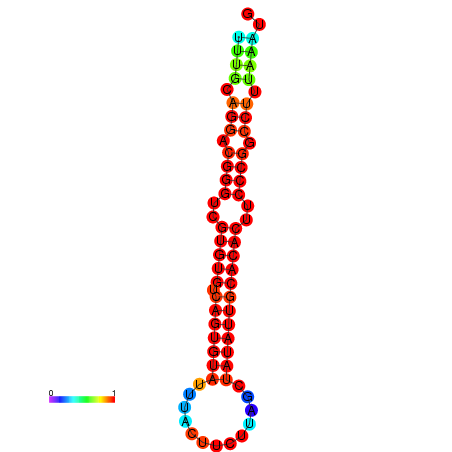 |
| dG=-24.8, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|
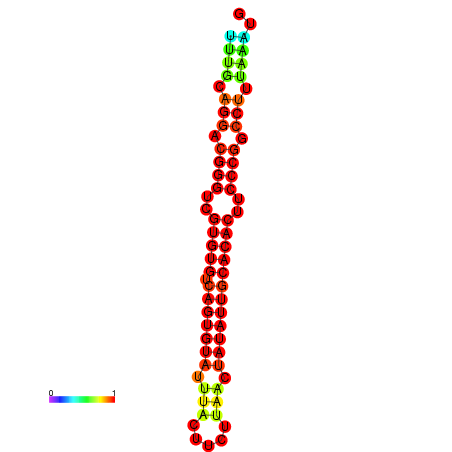 |
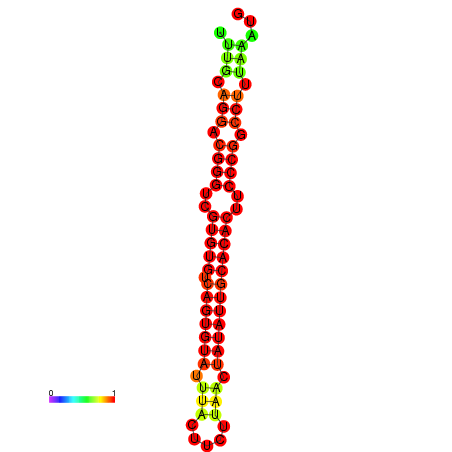 |
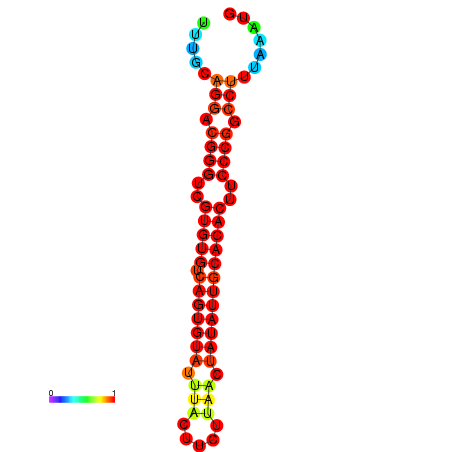 |
| dG=-25.2, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|---|---|---|
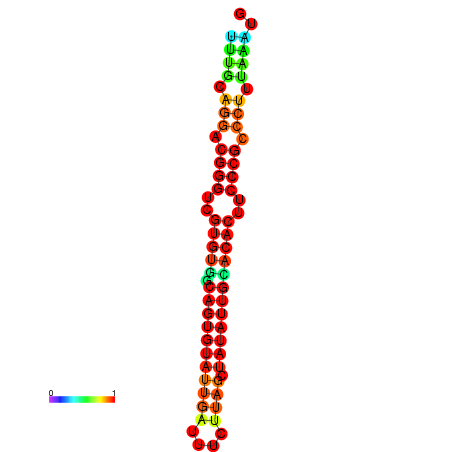 |
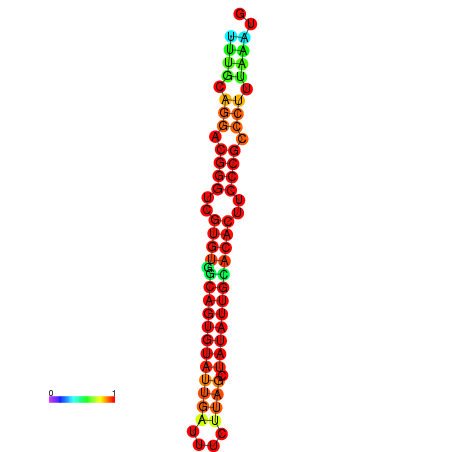 |
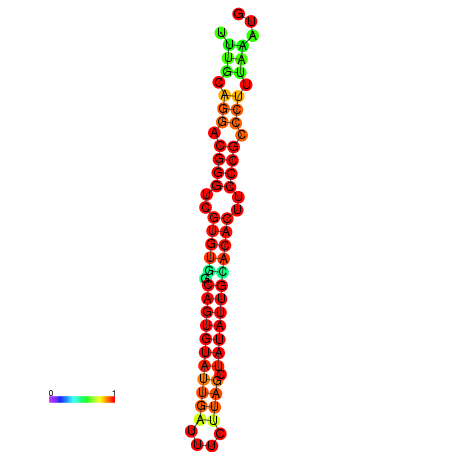 |
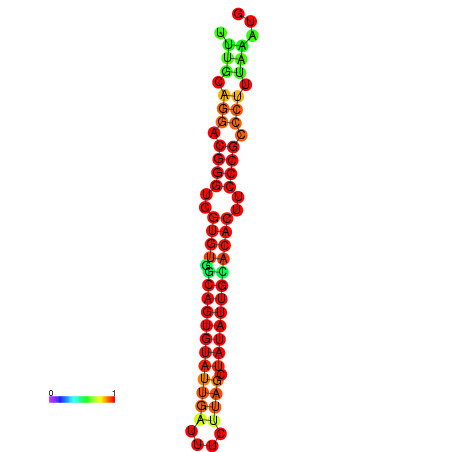 |
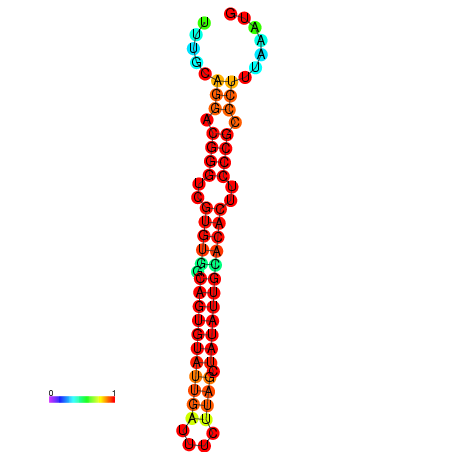 |
| dG=-25.2, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.3, p-value=0.009901 | dG=-24.2, p-value=0.009901 |
|---|---|---|---|---|
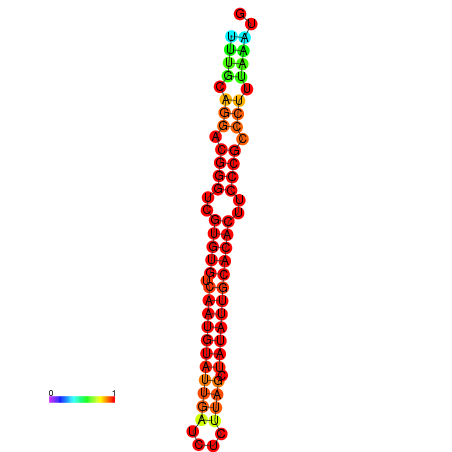 |
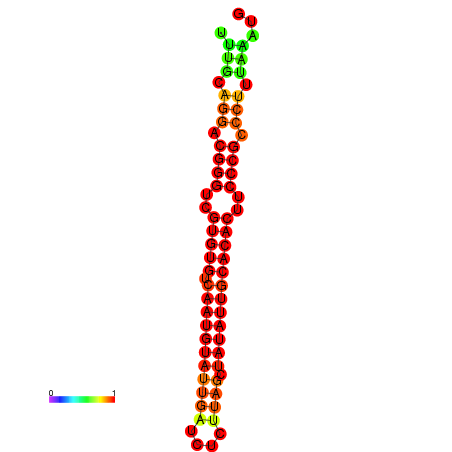 |
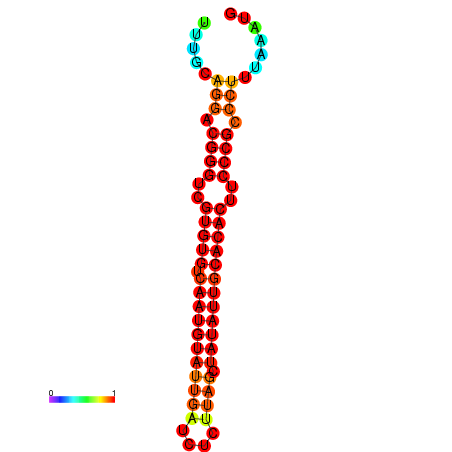 |
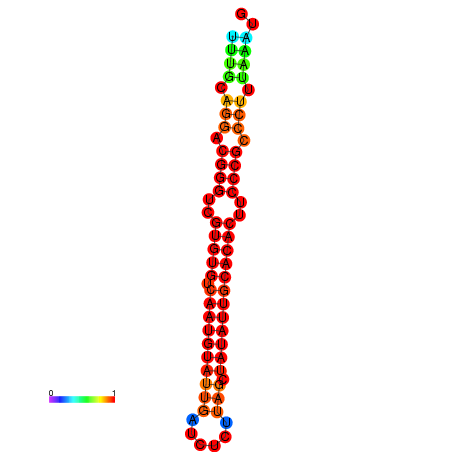 |
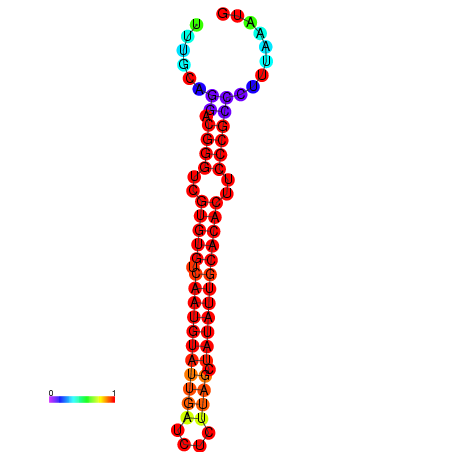 |
| dG=-22.3, p-value=0.009901 | dG=-21.9, p-value=0.009901 | dG=-21.9, p-value=0.009901 | dG=-21.9, p-value=0.009901 | dG=-21.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
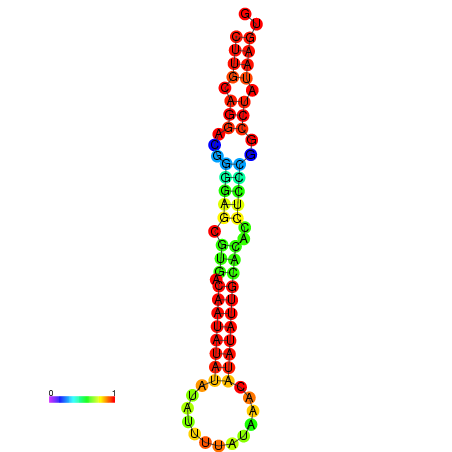 |
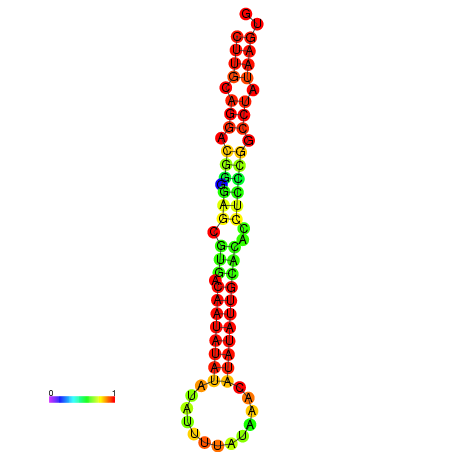
| dG=-17.5, p-value=0.009901 | dG=-17.2, p-value=0.009901 |
|---|---|
 |
 |
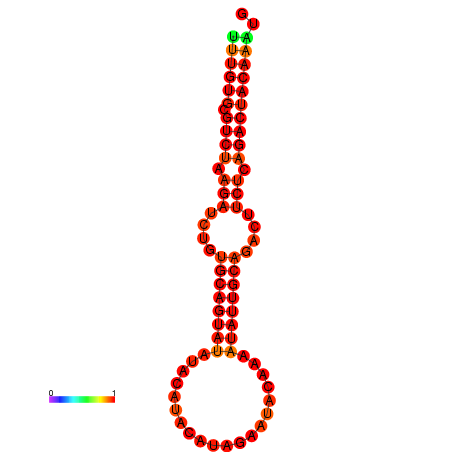
| dG=-17.9, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.2, p-value=0.009901 | dG=-16.9, p-value=0.009901 |
|---|---|---|---|
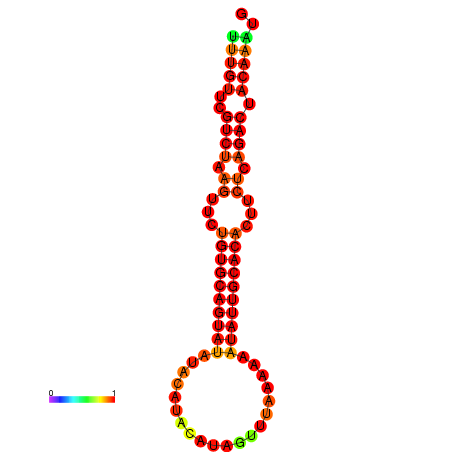 |
 |
 |
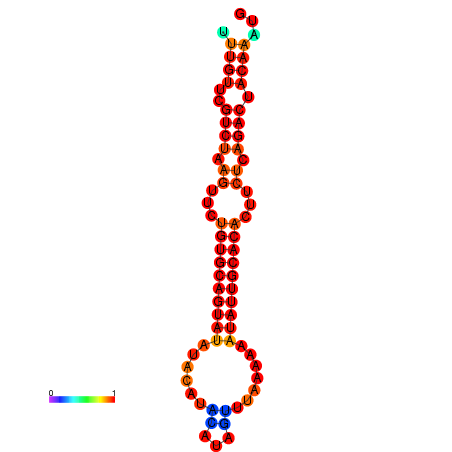 |
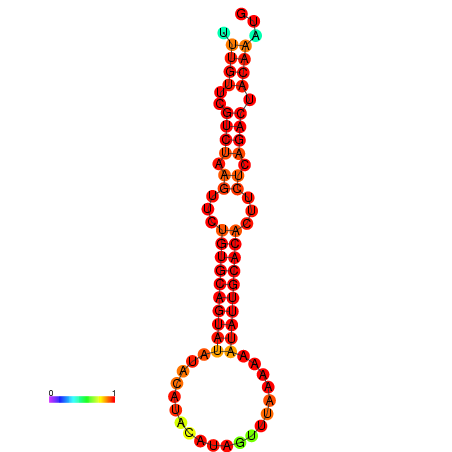
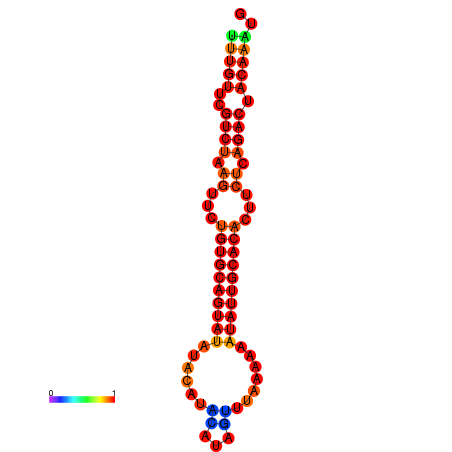
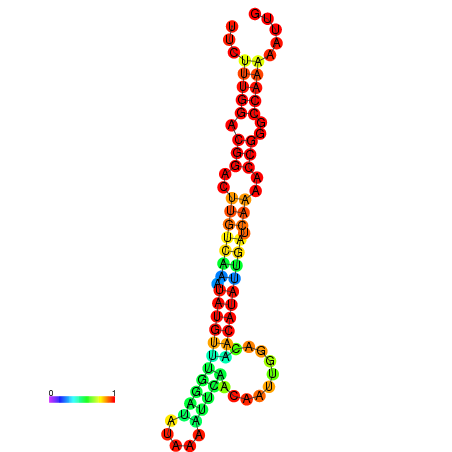
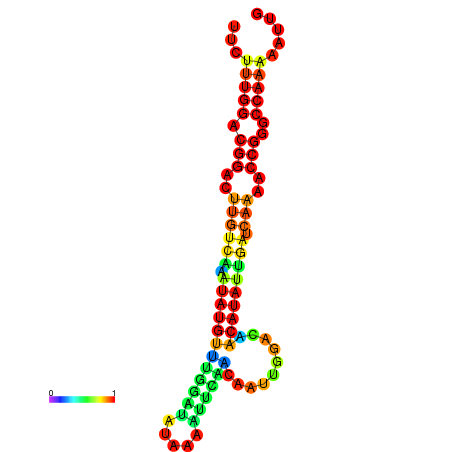
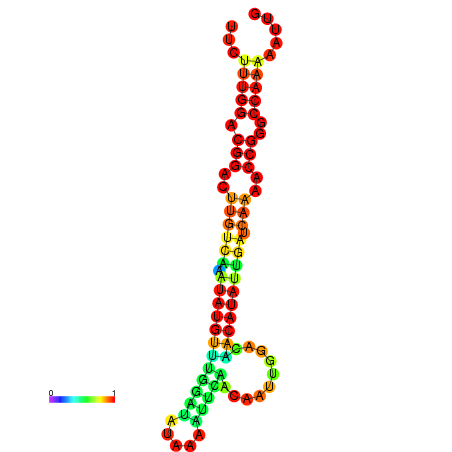
| dG=-16.6, p-value=0.009901 | dG=-16.6, p-value=0.009901 | dG=-16.6, p-value=0.009901 | dG=-16.6, p-value=0.009901 | dG=-16.6, p-value=0.009901 |
|---|---|---|---|---|
 |
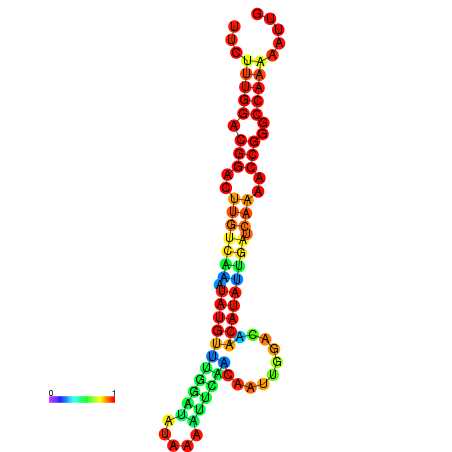 |
 |
 |
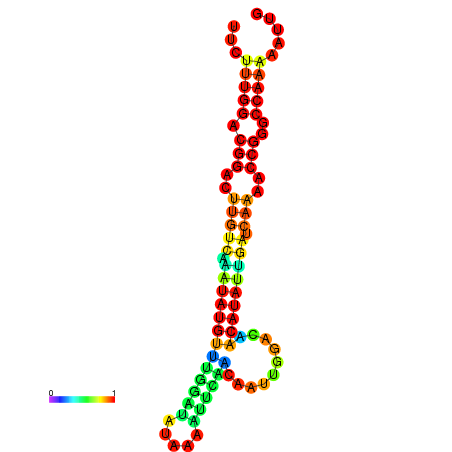 |
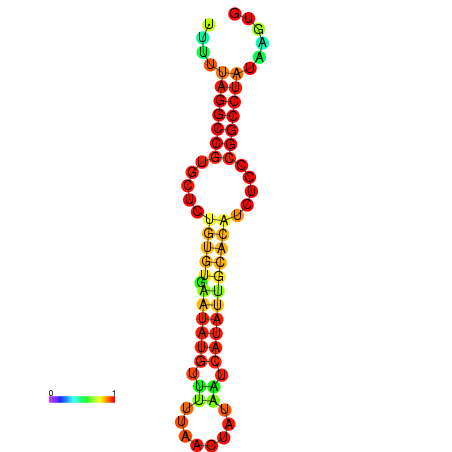
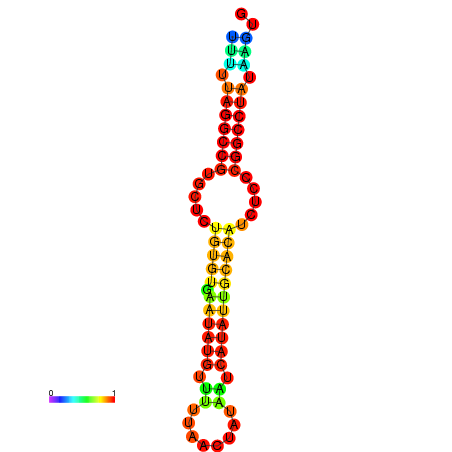
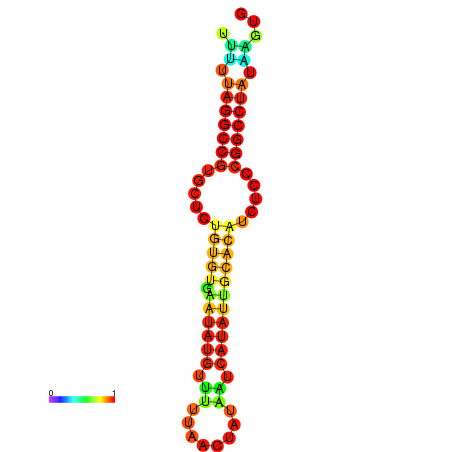
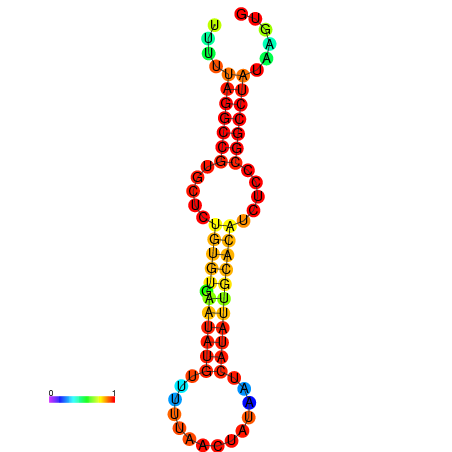
| dG=-20.7, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
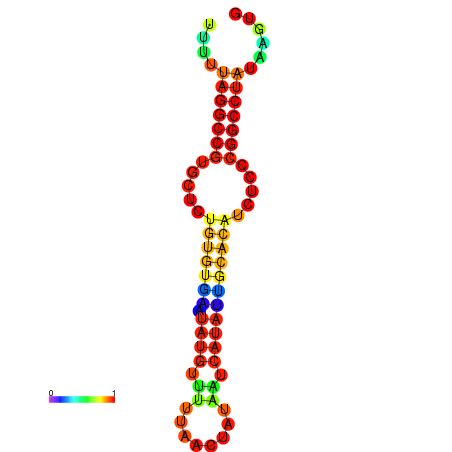 |
| dG=-23.6, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-22.7, p-value=0.009901 | dG=-22.7, p-value=0.009901 | dG=-22.7, p-value=0.009901 |
|---|---|---|---|---|
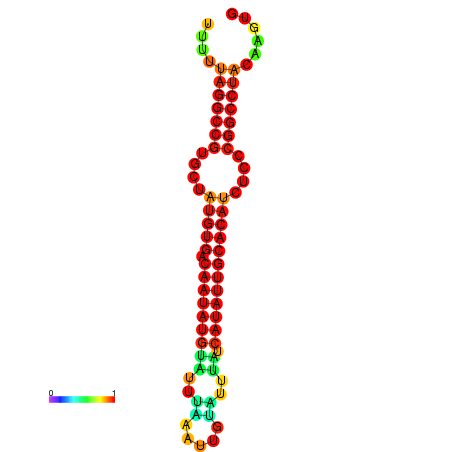 |
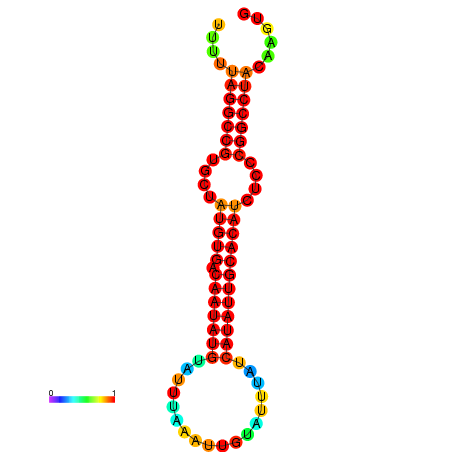 |
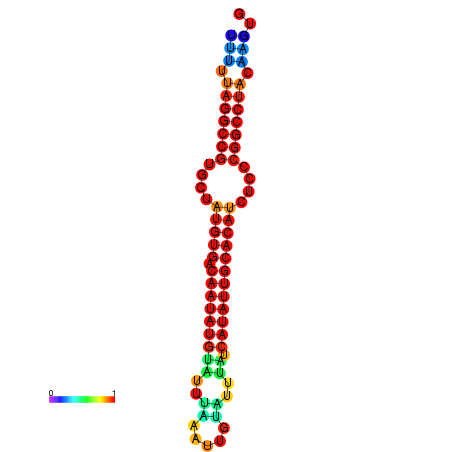 |
 |
 |
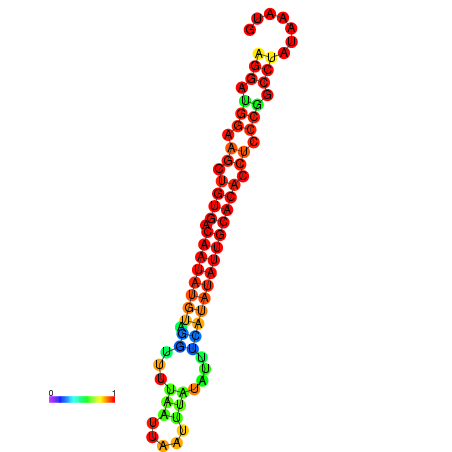
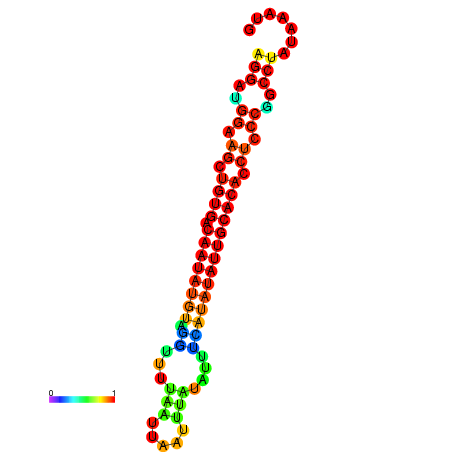
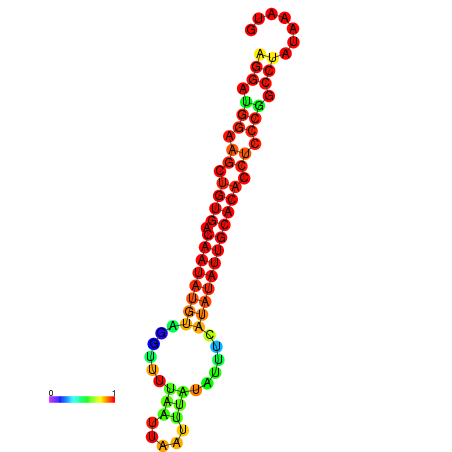
| dG=-18.2, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.0, p-value=0.009901 | dG=-17.9, p-value=0.009901 | dG=-17.7, p-value=0.009901 |
|---|---|---|---|---|
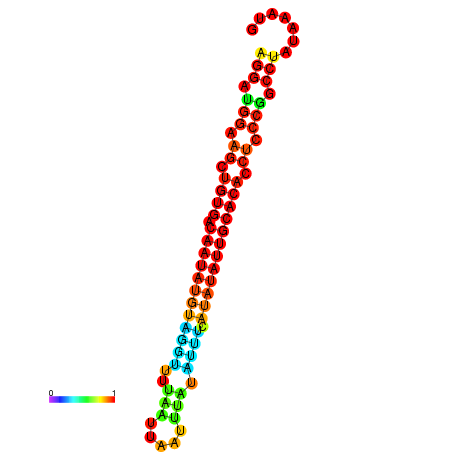 |
 |
 |
 |
 |
Generated: 03/07/2013 at 05:15 PM