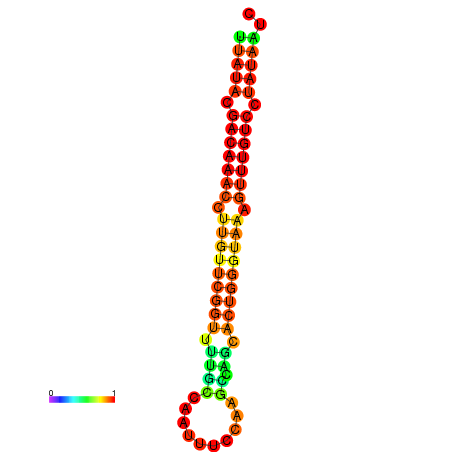
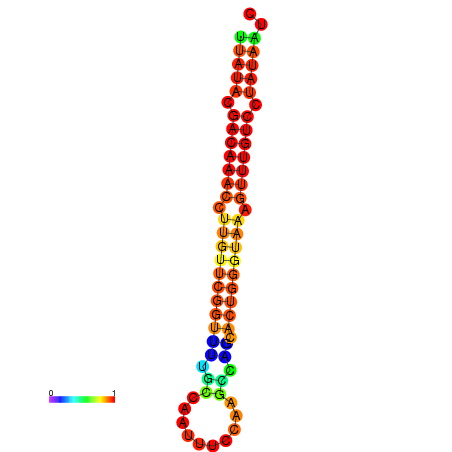



| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15549196-15549294 - | AA-------GTTT---GGAATAG-CAATTATACGACAAACCTTGTTCGGTTTTGCCA----------------ATTTCCA------AG-CCAGCACTGGGTAAAGTTTGTCCTATAATCCCGAAACGA--------------C-GACC |
| droSim1 | chr2R:14254629-14254727 - | AA-------GTTT---GGAACAG-CAATTATACGACAAACCTTGTTCGGTTTTGCCA----------------ATTTCCA------AG-CCAGCACTGGGTAAAGTTTGTCCTATAATCCCGAAACGA--------------C-GACC |
| droSec1 | super_1:13063923-13064021 - | AA-------GTTT---GGAACAG-CAATTATACGACAAACCTTGTTCGGTTTTGCCA----------------ATTTCCA------AG-CCAGCACTGGGTAAAGTTTGTCCTATAATCCCGAAACGA--------------C-GACC |
| droYak2 | chr2R:13623487-13623590 + | AA--GCAAAGTTT---GGAACAG-CAATTATACGACAAACCTTGTTCTGTTTTGCCA----------------ATTTCCT------AG-CCAGCACTGGGTAAAGTTTGTCCTATAATCCCGAAACGA--------------C-GACC |
| droEre2 | scaffold_4845:9749440-9749538 - | GA-------GTTT---GGAACAG-CAATTATACGACAAACCTTGTTCGGTTTTGCCA----------------ATTTCCT------AG-CCAGCACTGGGTAAAGTTTGTCCTATAATCCCGAAACGA--------------C-GACC |
| droAna3 | scaffold_13266:9731105-9731197 + | GA-----------------AGAT-C-ATTATACGACAAACCTTGTTCGGTTTTGCCA----------------ATTTCCT------A--GCAGCACTGGGTAAAGTTTGTCCTATAATCTATAAAAAA--------G-----CCGTCC |
| dp4 | chr3:12677873-12677971 - | AA-------CTAT---CTCGGAA-CTATTATACGATAAACTTTGTTCAGTTTTGCCA----------------ACTTGCT------T--GCAGCACTGGGTGAAGTTTGTCTTATAATCCCACAGTAA--------G-----C-GATC |
| droPer1 | super_2:8905688-8905793 + | AAAAACGAACTATCTCGGA---A-CTATTATACGATAAACTTTGTTCAGTTTTGCCA----------------ACTTGCT------T--GCAGCACTGGGTGAAGTTTGTCTTATAATCCCACAGTAA--------G-----C-GATC |
| droWil1 | scaffold_180701:1652063-1652174 - | A---------TTT---CATATAA-T-ATTATATGATAAACTTTGTTCAGTTTTGCAATCAAAAAATCAAGAAAA-TTCCT------TA-GCATCACTGGGTGAAGTTTGTCTTATAATCTTTAAACTA--------------C-AATC |
| droVir3 | scaffold_12875:4111798-4111897 + | TA---------------TAACAA-ATATTATATGGTAAACTTTGTTCAGTTTTGCCA----------------GGCTCTT-TTTGTGC-GCATCACTGGGTGAAGTTTGTCGTATAATCGCAAAAAGA--------------A-CAGC |
| droMoj3 | scaffold_6496:20937092-20937209 - | TA-------TACA---AGAACAGAATATTATAAGGTAAACTTTGTTCAGTTTTGCCG----------------GGTTCTATGTTGTGC-GCATCACTGGGTGAAGTTTGTCGTATAATCG--AAACGAAACAGCAAGGAAAAA-GTCC |
| droGri2 | scaffold_15112:2109131-2109231 - | AA-------ATAT---ATAATTA-C-ATTATATGGCAAACCTTGTTCAGTTTTGACA----------------ACTTTGTTTCTGTGCATCATCACTGGGTGAAGTTTGTCGTATAATC--GAAATG-----------------GATC |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15549196-15549294 - | AA-------GTTT---GGAATAG-CAATTATACGACAAACCTTGTTCGGTTTTGCCA----------------ATTTCCA------AG-CCAGCACTGGGTAAAGTTTGTCCTATAATCCCGAAACGA--------------C-GACC |
| droSim1 | chr2R:14254629-14254727 - | AA-------GTTT---GGAACAG-CAATTATACGACAAACCTTGTTCGGTTTTGCCA----------------ATTTCCA------AG-CCAGCACTGGGTAAAGTTTGTCCTATAATCCCGAAACGA--------------C-GACC |
| droSec1 | super_1:13063923-13064021 - | AA-------GTTT---GGAACAG-CAATTATACGACAAACCTTGTTCGGTTTTGCCA----------------ATTTCCA------AG-CCAGCACTGGGTAAAGTTTGTCCTATAATCCCGAAACGA--------------C-GACC |
| droYak2 | chr2R:13623487-13623590 + | AA--GCAAAGTTT---GGAACAG-CAATTATACGACAAACCTTGTTCTGTTTTGCCA----------------ATTTCCT------AG-CCAGCACTGGGTAAAGTTTGTCCTATAATCCCGAAACGA--------------C-GACC |
| droEre2 | scaffold_4845:9749440-9749538 - | GA-------GTTT---GGAACAG-CAATTATACGACAAACCTTGTTCGGTTTTGCCA----------------ATTTCCT------AG-CCAGCACTGGGTAAAGTTTGTCCTATAATCCCGAAACGA--------------C-GACC |
| droAna3 | scaffold_13266:9731105-9731197 + | GA-----------------AGAT-C-ATTATACGACAAACCTTGTTCGGTTTTGCCA----------------ATTTCCT------A--GCAGCACTGGGTAAAGTTTGTCCTATAATCTATAAAAAA--------G-----CCGTCC |
| dp4 | chr3:12677873-12677971 - | AA-------CTAT---CTCGGAA-CTATTATACGATAAACTTTGTTCAGTTTTGCCA----------------ACTTGCT------T--GCAGCACTGGGTGAAGTTTGTCTTATAATCCCACAGTAA--------G-----C-GATC |
| droPer1 | super_2:8905688-8905793 + | AAAAACGAACTATCTCGGA---A-CTATTATACGATAAACTTTGTTCAGTTTTGCCA----------------ACTTGCT------T--GCAGCACTGGGTGAAGTTTGTCTTATAATCCCACAGTAA--------G-----C-GATC |
| droWil1 | scaffold_180701:1652063-1652174 - | A---------TTT---CATATAA-T-ATTATATGATAAACTTTGTTCAGTTTTGCAATCAAAAAATCAAGAAAA-TTCCT------TA-GCATCACTGGGTGAAGTTTGTCTTATAATCTTTAAACTA--------------C-AATC |
| droVir3 | scaffold_12875:4111798-4111897 + | TA---------------TAACAA-ATATTATATGGTAAACTTTGTTCAGTTTTGCCA----------------GGCTCTT-TTTGTGC-GCATCACTGGGTGAAGTTTGTCGTATAATCGCAAAAAGA--------------A-CAGC |
| droMoj3 | scaffold_6496:20937092-20937209 - | TA-------TACA---AGAACAGAATATTATAAGGTAAACTTTGTTCAGTTTTGCCG----------------GGTTCTATGTTGTGC-GCATCACTGGGTGAAGTTTGTCGTATAATCG--AAACGAAACAGCAAGGAAAAA-GTCC |
| droGri2 | scaffold_15112:2109131-2109231 - | AA-------ATAT---ATAATTA-C-ATTATATGGCAAACCTTGTTCAGTTTTGACA----------------ACTTTGTTTCTGTGCATCATCACTGGGTGAAGTTTGTCGTATAATC--GAAATG-----------------GATC |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-15.7, p-value=0.009901 | dG=-15.6, p-value=0.009901 | dG=-15.5, p-value=0.009901 | dG=-15.4, p-value=0.009901 | dG=-15.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-15.7, p-value=0.009901 | dG=-15.6, p-value=0.009901 | dG=-15.5, p-value=0.009901 | dG=-15.4, p-value=0.009901 | dG=-15.3, p-value=0.009901 |
|---|---|---|---|---|
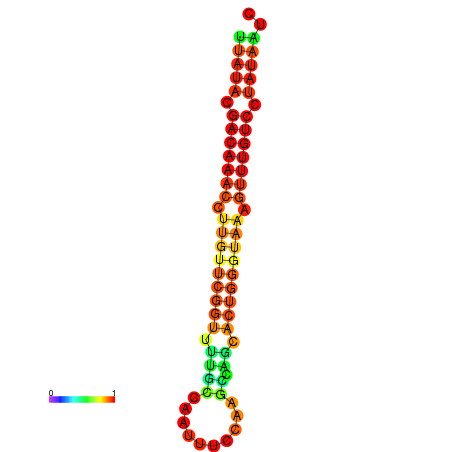 |
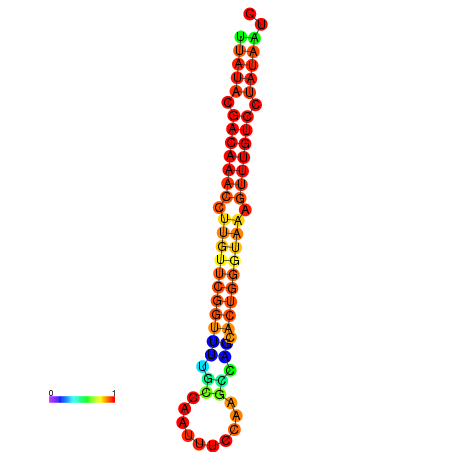 |
 |
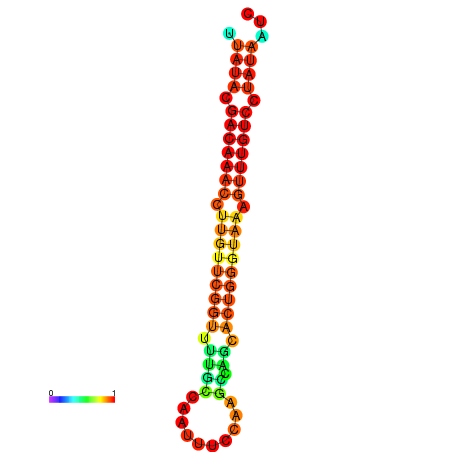 |
 |
| dG=-15.7, p-value=0.009901 | dG=-15.6, p-value=0.009901 | dG=-15.5, p-value=0.009901 | dG=-15.4, p-value=0.009901 | dG=-15.3, p-value=0.009901 |
|---|---|---|---|---|
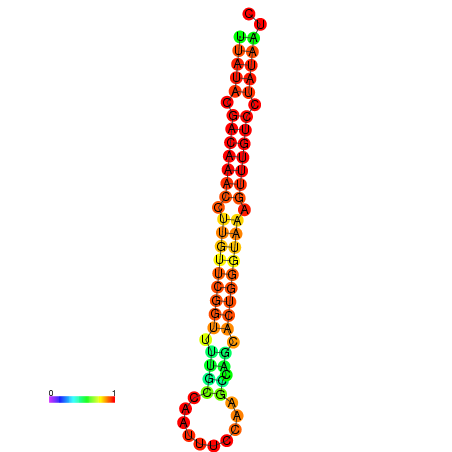 |
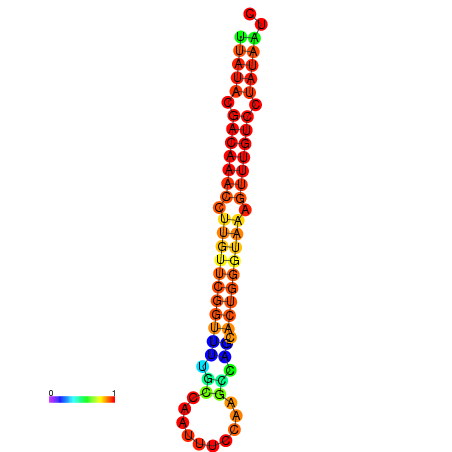 |
 |
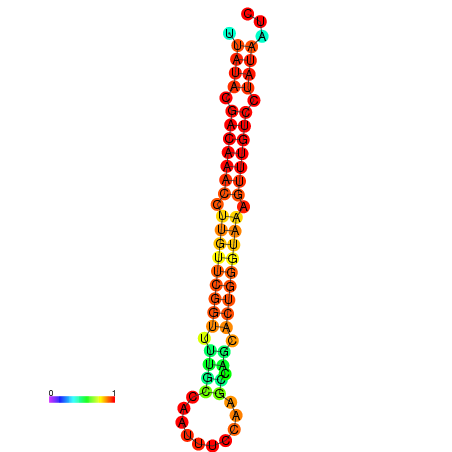 |
 |
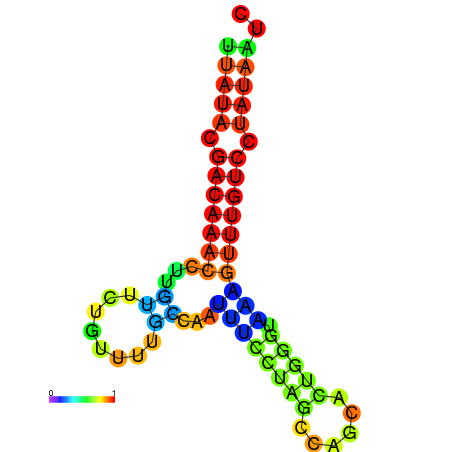
| dG=-12.9, p-value=0.009901 | dG=-12.7, p-value=0.009901 | dG=-12.6, p-value=0.009901 | dG=-12.6, p-value=0.009901 | dG=-12.4, p-value=0.009901 |
|---|---|---|---|---|
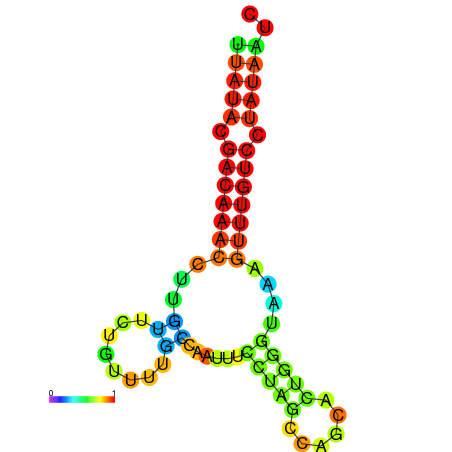 |
 |
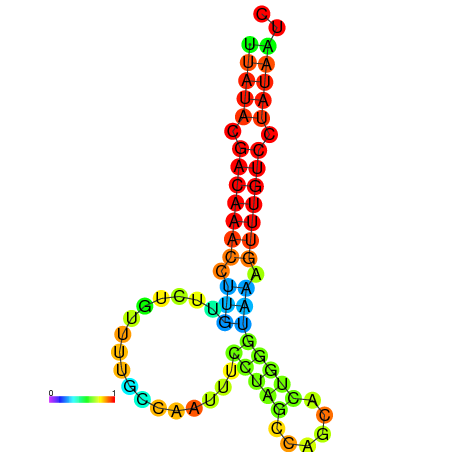 |
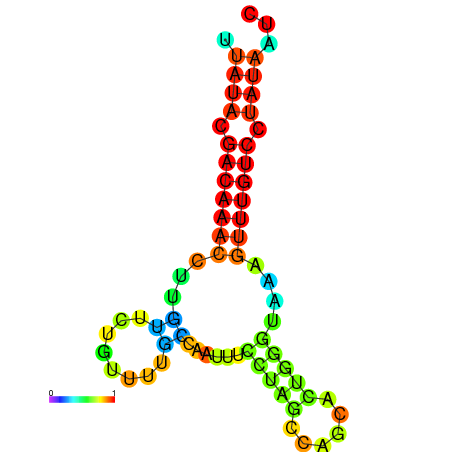 |
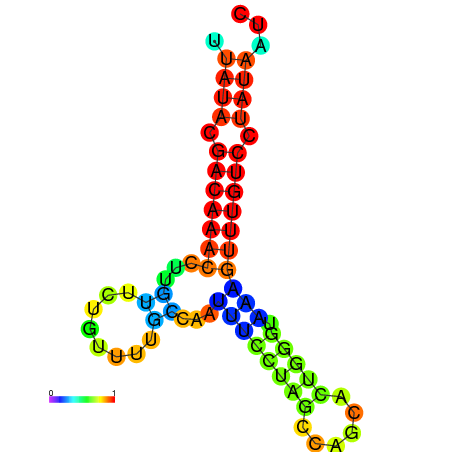 |
| dG=-15.7, p-value=0.009901 | dG=-15.6, p-value=0.009901 | dG=-15.5, p-value=0.009901 | dG=-15.4, p-value=0.009901 | dG=-15.3, p-value=0.009901 |
|---|---|---|---|---|
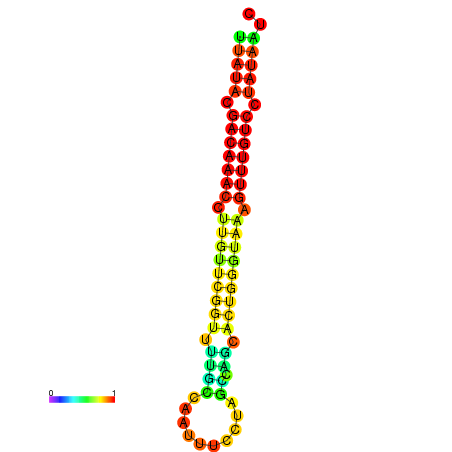 |
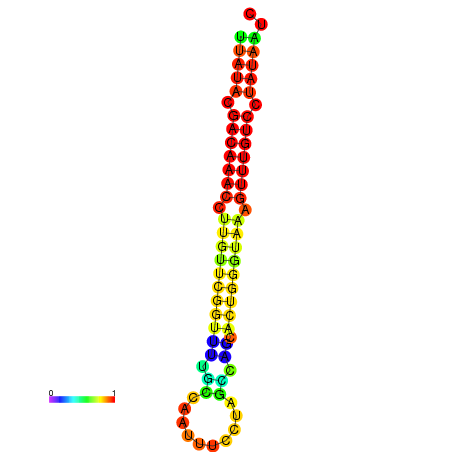 |
 |
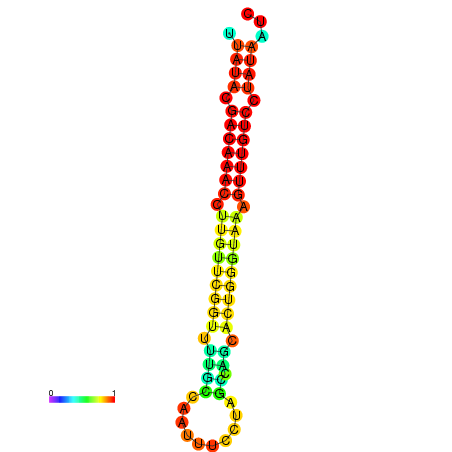 |
 |
| dG=-19.5, p-value=0.009901 | dG=-19.2, p-value=0.009901 |
|---|---|
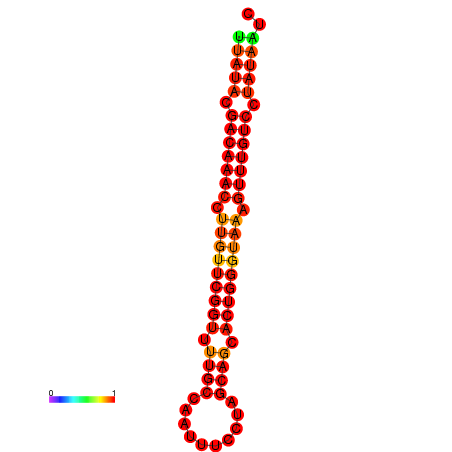 |
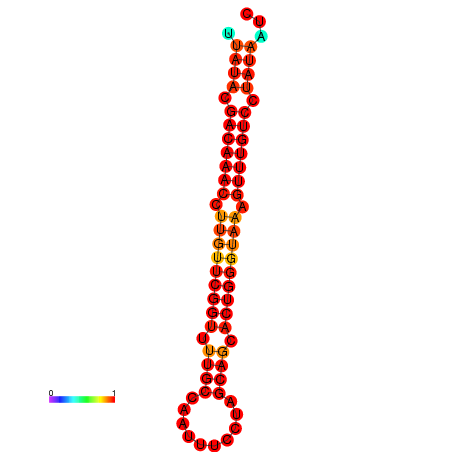 |
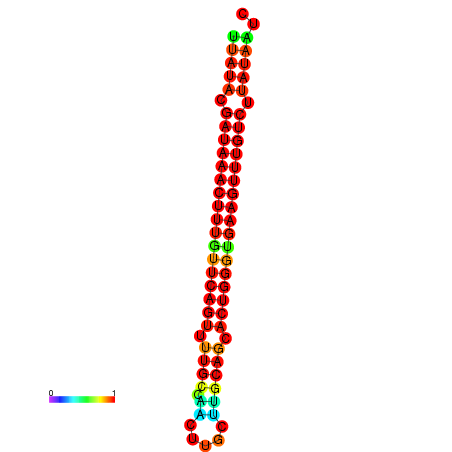
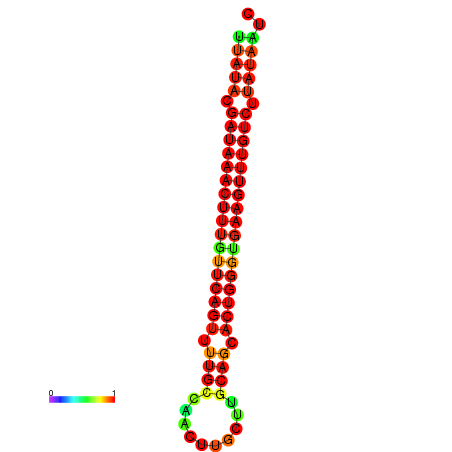
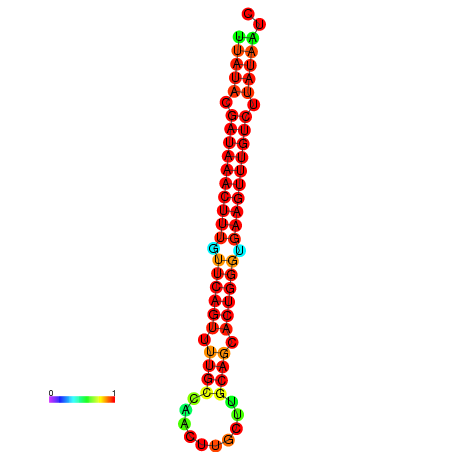
| dG=-21.4, p-value=0.009901 | dG=-21.1, p-value=0.009901 | dG=-21.1, p-value=0.009901 | dG=-20.8, p-value=0.009901 | dG=-20.8, p-value=0.009901 |
|---|---|---|---|---|
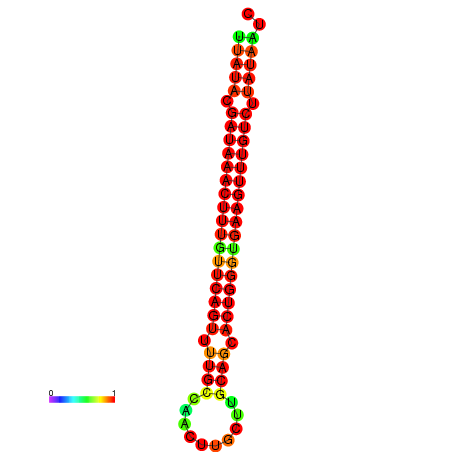 |
 |
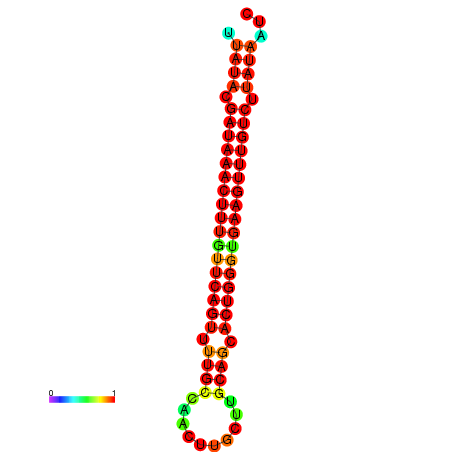 |
 |
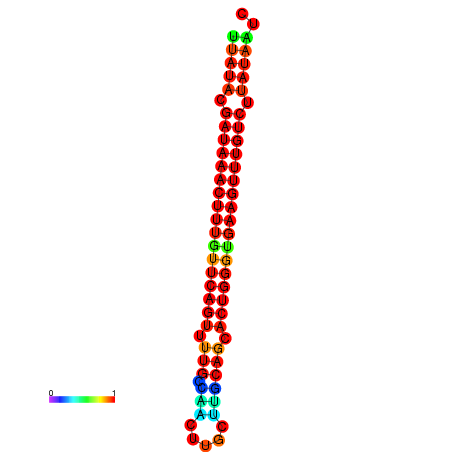 |
| dG=-21.4, p-value=0.009901 | dG=-21.1, p-value=0.009901 | dG=-21.1, p-value=0.009901 | dG=-20.8, p-value=0.009901 | dG=-20.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
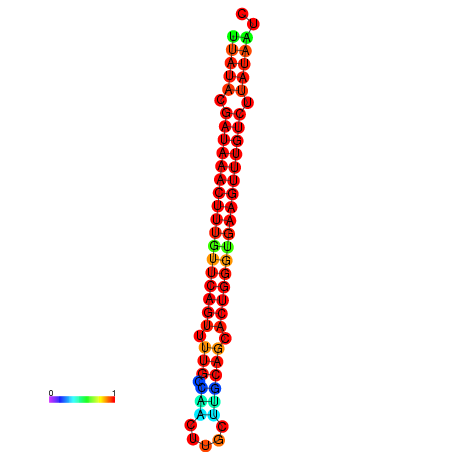 |
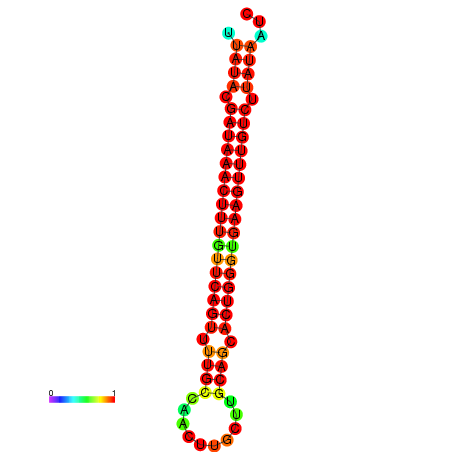
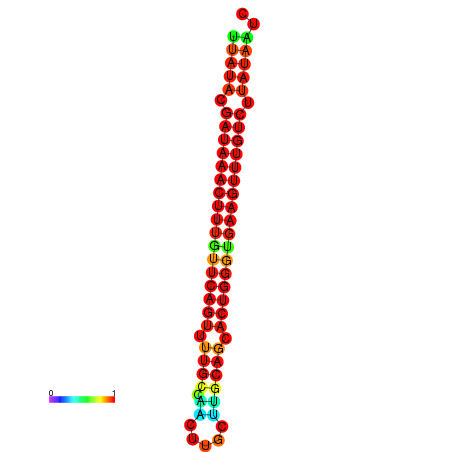
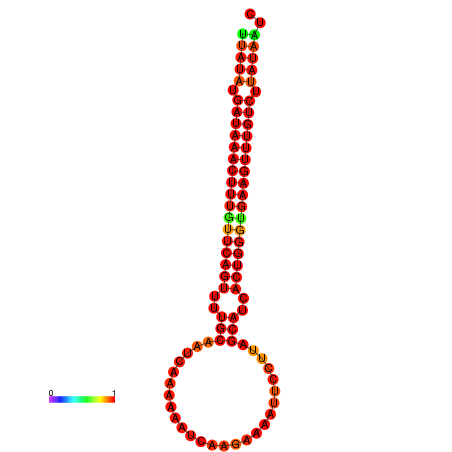
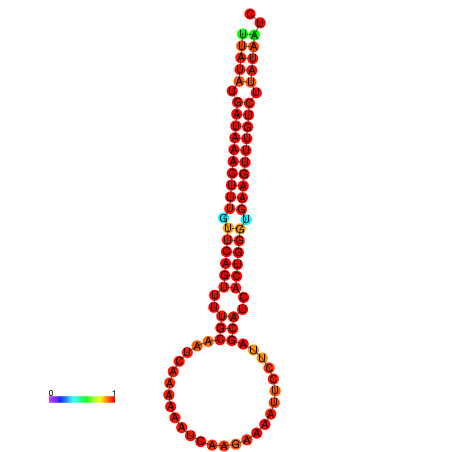
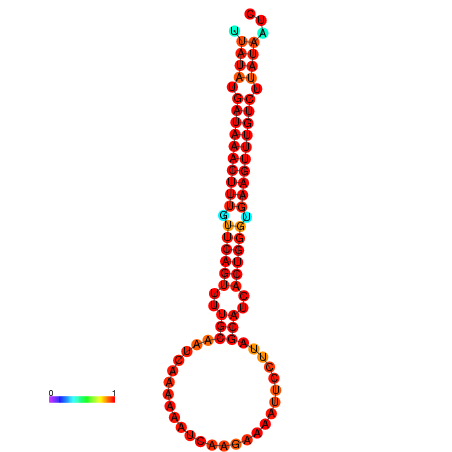
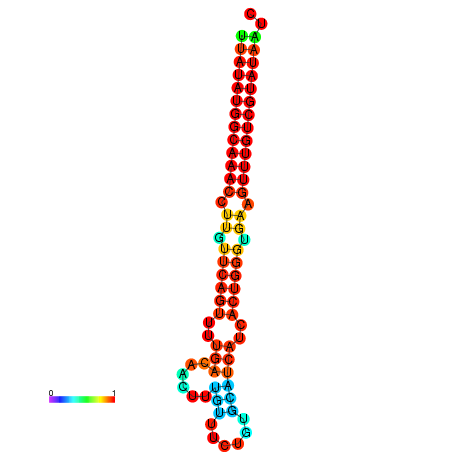
| dG=-21.1, p-value=0.009901 | dG=-20.8, p-value=0.009901 | dG=-20.8, p-value=0.009901 | dG=-20.5, p-value=0.009901 |
|---|---|---|---|
 |
 |
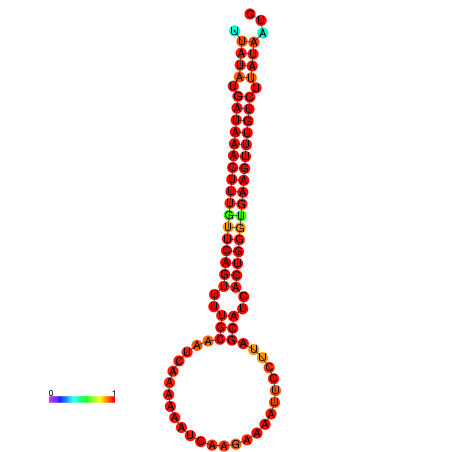 |
 |
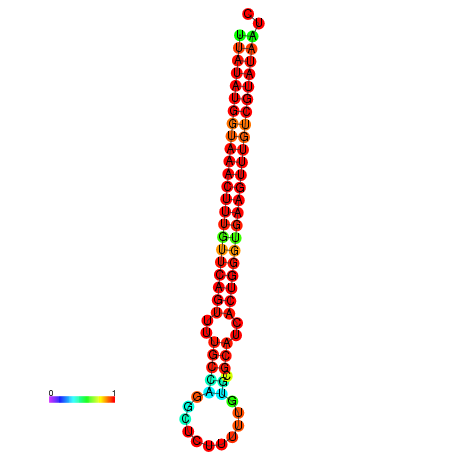
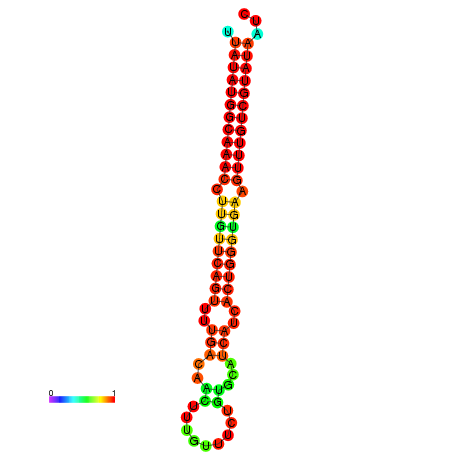
| dG=-23.2, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.9, p-value=0.009901 | dG=-22.9, p-value=0.009901 |
|---|---|---|---|---|
 |
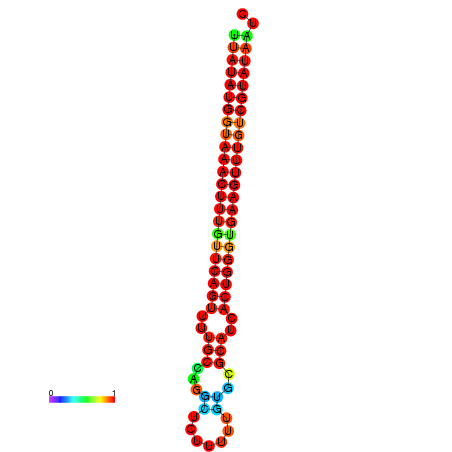 |
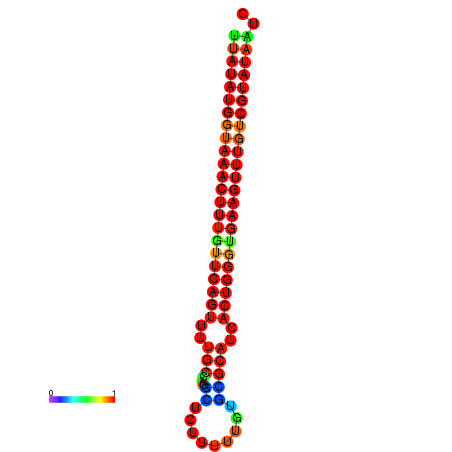 |
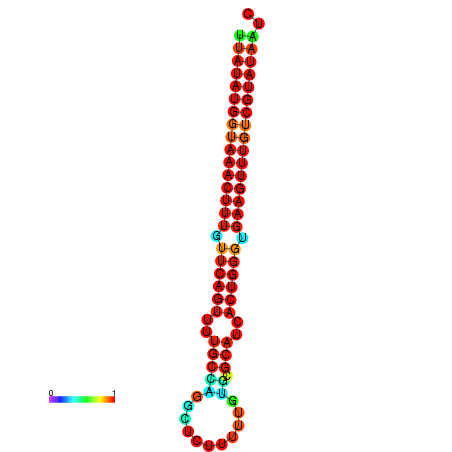 |
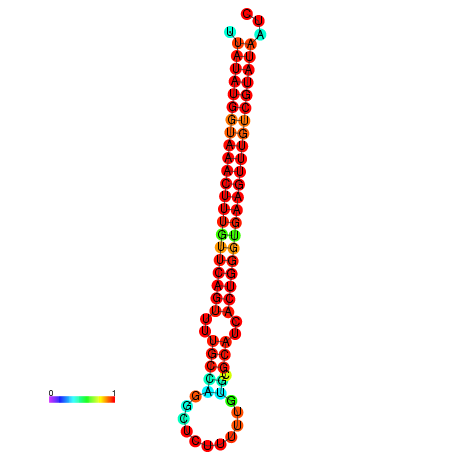 |
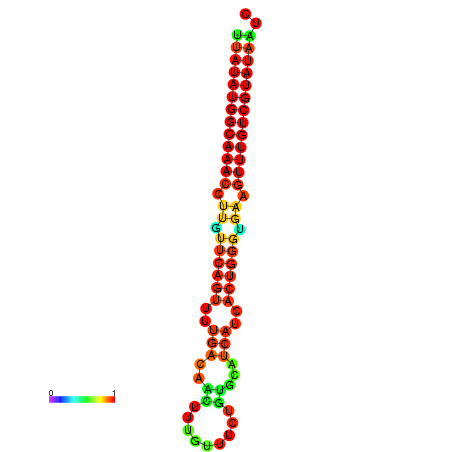
| dG=-18.4, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-17.8, p-value=0.009901 | dG=-17.4, p-value=0.009901 |
|---|---|---|---|---|
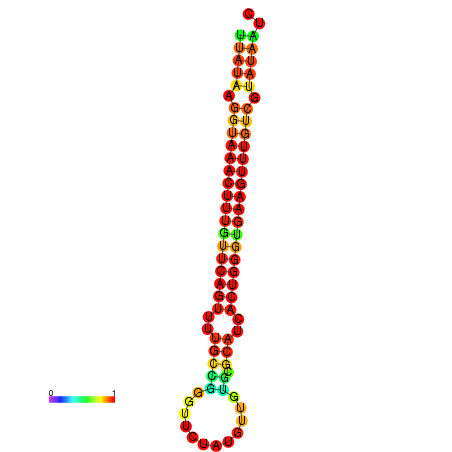 |
 |
 |
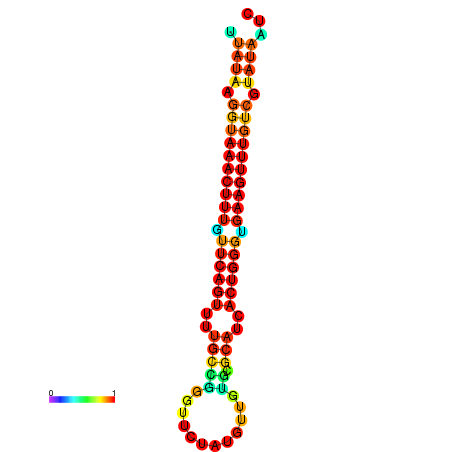 |
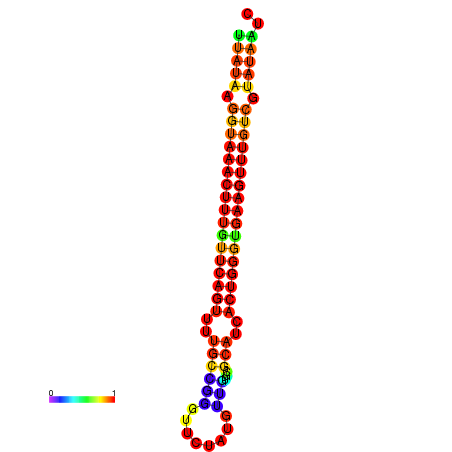 |
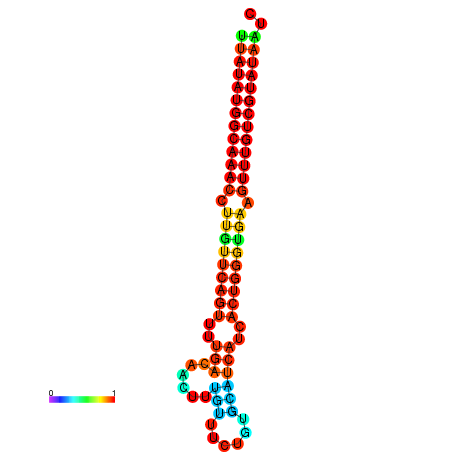
| dG=-21.8, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.2, p-value=0.009901 |
|---|---|---|---|---|
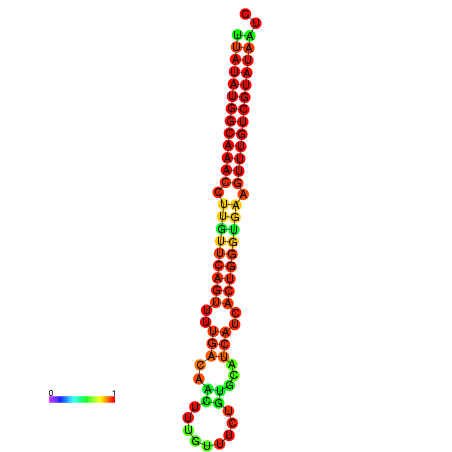 |
 |
 |
 |
 |
Generated: 03/07/2013 at 05:14 PM