

| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:10103292-10103384 - | AACACG--TCTCGACGCCTCGCAGTATATTTTTGTGTTTTGTTTCGTTTTGCAATCCAAATCACAGGATTATACTGTGAGATGACCAGCGTG---CCA |
| droSim1 | chr2R:8866656-8866748 - | CACACG--TCTCGACGCCTCGCAGTATATTTTTGTGTTTTGTTTCGTTTTGCAACTCAAATCACAGGATTATACTGTGAGATGACCAGCGTG---CCA |
| droSec1 | super_1:7611688-7611779 - | CA--CG--TCTCGACGCCTCGCAGTATATTTTTGTGTTTTGTTTCGTTTTGCAACTCAAATCACAGGATTATACTGTGAGATGACCAGCGTG---CCA |
| droYak2 | chr2R:10041132-10041224 - | CACACG--TCTCGACGCCTCGCAGTATATTTTTGTGTTTTGTTTCCTTTTGCAACTCAAATCACAGGATTATACTGTGAGATGACCAGCGTG---CCA |
| droEre2 | scaffold_4845:15739235-15739330 + | CACACG--TCTCGACGCCTCGCAGTATATTTTTGTGTTTTGTTTCGTTTTGCAAATCAAATCACAGGATTATACTGTGAGATGACCAGCGTGCCGCCA |
| droAna3 | scaffold_13266:5819955-5820045 + | CA--TG--TCTCGTCGCCTCGCAGTATATTTTTGTGTTTTGTTTGG-TTTGAAACTCAAATCACAGGATTATACTGTGAGATGCCAAGCGTG---TCA |
| dp4 | chr3:16336992-16337078 - | --------TCTTGACGCCTCGCAGTATATTTTTGTGTTTTGTTGTG-TCTGAAACGCAAATCACAGGATTATACTGTGAGATGCTGGGCGTG---CTA |
| droPer1 | super_4:426256-426345 + | G---CG--TCTTGACGCCTCGCAGTATATTTTTGTGTTTTGTTGTG-TCTGAAACGCAAATCACAGGATTATACTGTGAGATGCTGGGCGTG---CTA |
| droWil1 | scaffold_180700:1860929-1861010 + | AGACTA--TTCTTGCGTCTCGCAGTATATTTTTGTGTTTTGTTCAATTTGTCAACTCAAATCACAGGATTATACTGTGAGCTGC-------------- |
| droVir3 | scaffold_12875:17605293-17605383 + | AACGCA--T-TCGATGTCTCGCAGTATACTTTTGTGTTTTGTTTAT-TTTGAAAATCAAATCACAGGATTATACTGTGAGTTATCGGCTGTA---GCA |
| droMoj3 | scaffold_6496:11229058-11229148 + | AACACA--T-TCGATGTCTCGCAGTATATTTTTGTGTTTTGTTTAT-TATAAAAATCAAATCACAGGATTATACTGTGAGTTATCGGCTGAA---ACA |
| droGri2 | scaffold_15245:14076242-14076330 - | CCCACCACTGTCGATGTCTCGCAGTATATTTTTGTGTTTTGTTTATTTTTGAAAATCAAATCACAGGATTATACTGTGAGTTATC------G---CCA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:10103292-10103384 - | AACACG--TCTCGACGCCTCGCAGTATATTTTTGTGTTTTGTTTCGTTTTGCAATCCAAATCACAGGATTATACTGTGAGATGACCAGCGTG---CCA |
| droSim1 | chr2R:8866656-8866748 - | CACACG--TCTCGACGCCTCGCAGTATATTTTTGTGTTTTGTTTCGTTTTGCAACTCAAATCACAGGATTATACTGTGAGATGACCAGCGTG---CCA |
| droSec1 | super_1:7611688-7611779 - | CA--CG--TCTCGACGCCTCGCAGTATATTTTTGTGTTTTGTTTCGTTTTGCAACTCAAATCACAGGATTATACTGTGAGATGACCAGCGTG---CCA |
| droYak2 | chr2R:10041132-10041224 - | CACACG--TCTCGACGCCTCGCAGTATATTTTTGTGTTTTGTTTCCTTTTGCAACTCAAATCACAGGATTATACTGTGAGATGACCAGCGTG---CCA |
| droEre2 | scaffold_4845:15739235-15739330 + | CACACG--TCTCGACGCCTCGCAGTATATTTTTGTGTTTTGTTTCGTTTTGCAAATCAAATCACAGGATTATACTGTGAGATGACCAGCGTGCCGCCA |
| droAna3 | scaffold_13266:5819955-5820045 + | CA--TG--TCTCGTCGCCTCGCAGTATATTTTTGTGTTTTGTTTGG-TTTGAAACTCAAATCACAGGATTATACTGTGAGATGCCAAGCGTG---TCA |
| dp4 | chr3:16336992-16337078 - | --------TCTTGACGCCTCGCAGTATATTTTTGTGTTTTGTTGTG-TCTGAAACGCAAATCACAGGATTATACTGTGAGATGCTGGGCGTG---CTA |
| droPer1 | super_4:426256-426345 + | G---CG--TCTTGACGCCTCGCAGTATATTTTTGTGTTTTGTTGTG-TCTGAAACGCAAATCACAGGATTATACTGTGAGATGCTGGGCGTG---CTA |
| droWil1 | scaffold_180700:1860929-1861010 + | AGACTA--TTCTTGCGTCTCGCAGTATATTTTTGTGTTTTGTTCAATTTGTCAACTCAAATCACAGGATTATACTGTGAGCTGC-------------- |
| droVir3 | scaffold_12875:17605293-17605383 + | AACGCA--T-TCGATGTCTCGCAGTATACTTTTGTGTTTTGTTTAT-TTTGAAAATCAAATCACAGGATTATACTGTGAGTTATCGGCTGTA---GCA |
| droMoj3 | scaffold_6496:11229058-11229148 + | AACACA--T-TCGATGTCTCGCAGTATATTTTTGTGTTTTGTTTAT-TATAAAAATCAAATCACAGGATTATACTGTGAGTTATCGGCTGAA---ACA |
| droGri2 | scaffold_15245:14076242-14076330 - | CCCACCACTGTCGATGTCTCGCAGTATATTTTTGTGTTTTGTTTATTTTTGAAAATCAAATCACAGGATTATACTGTGAGTTATC------G---CCA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-25.4, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-26.0, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.1, p-value=0.009901 |
|---|---|---|
 |
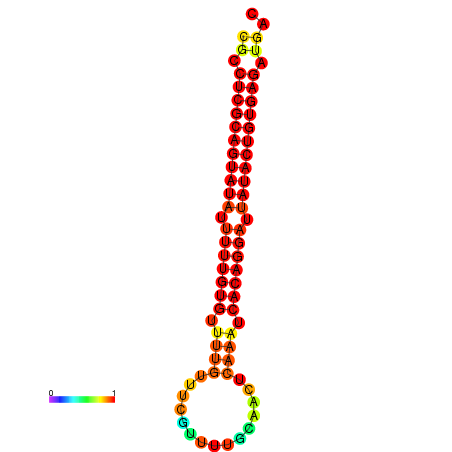 |
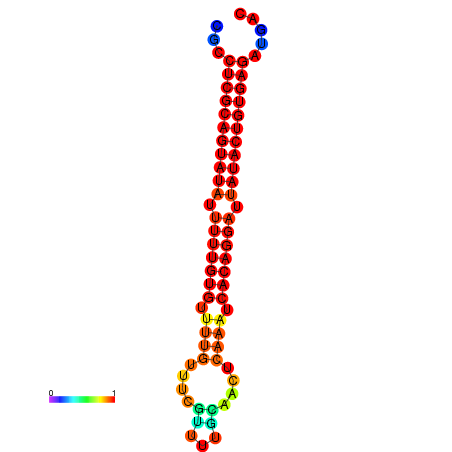 |
| dG=-26.0, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.1, p-value=0.009901 |
|---|---|---|
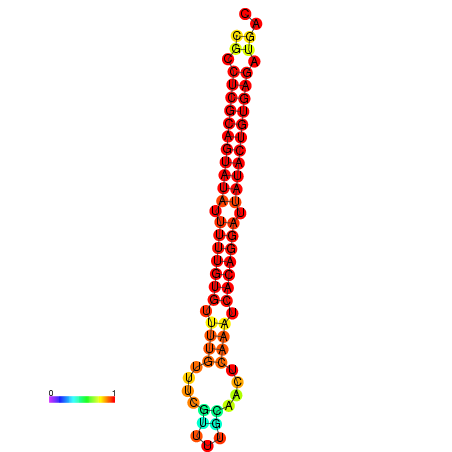 |
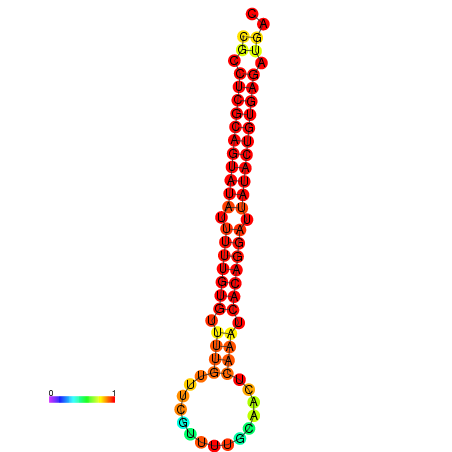 |
 |
| dG=-25.8, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|
 |
 |
| dG=-26.6, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
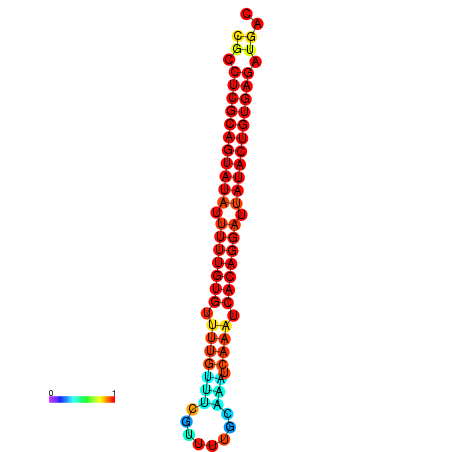 |
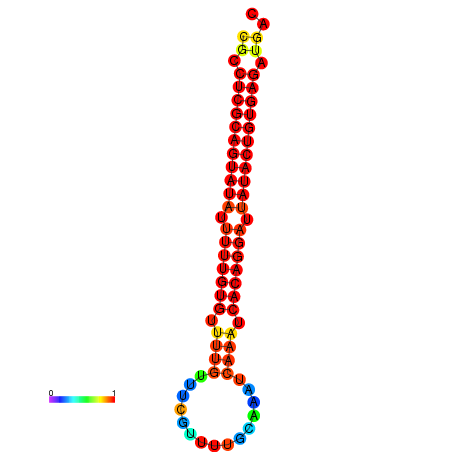 |
| dG=-26.1, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|---|---|
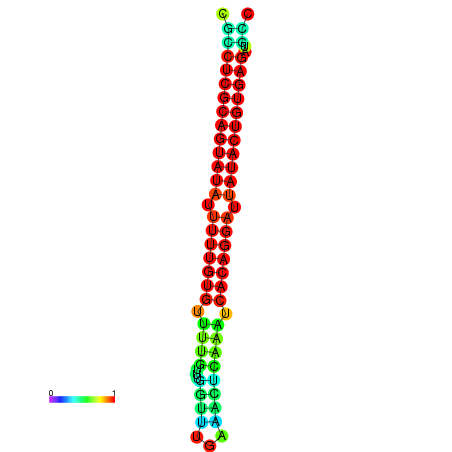 |
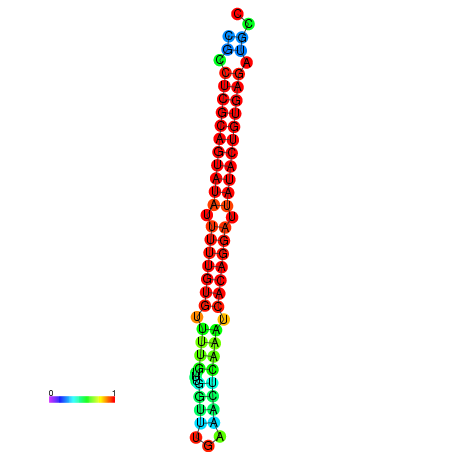 |
 |
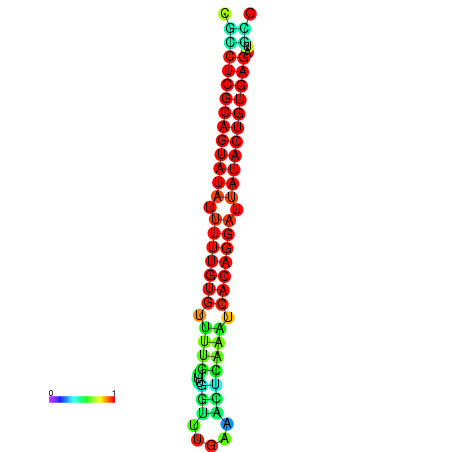 |
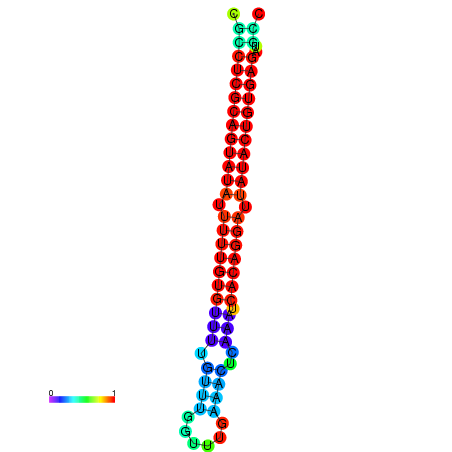 |
| dG=-28.1, p-value=0.009901 | dG=-28.1, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-28.0, p-value=0.009901 |
|---|---|---|---|---|
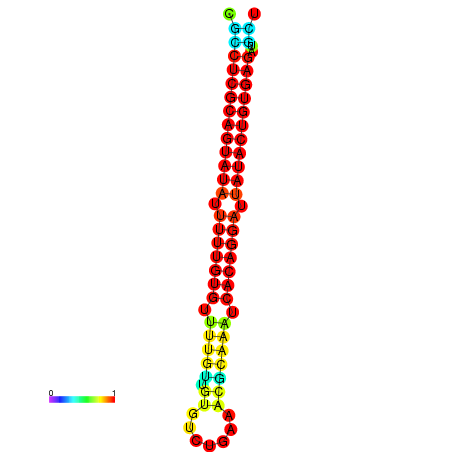 |
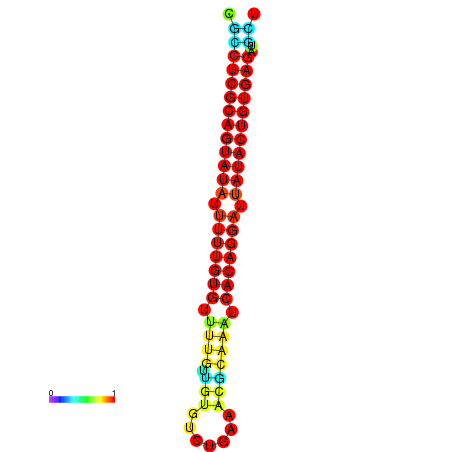 |
 |
 |
 |
| dG=-28.1, p-value=0.009901 | dG=-28.1, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-28.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-25.3, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|
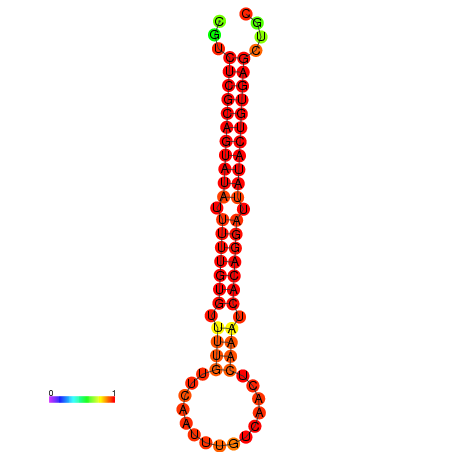 |
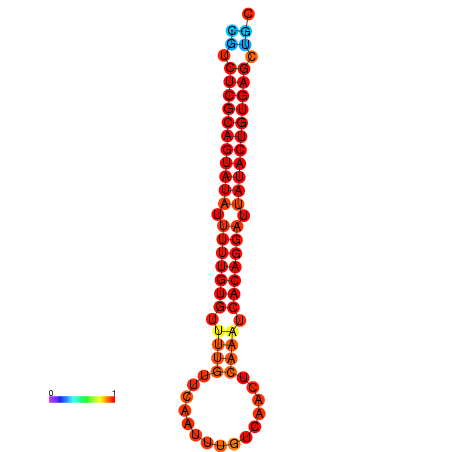 |
| dG=-24.8, p-value=0.009901 | dG=-24.4, p-value=0.009901 | dG=-24.4, p-value=0.009901 |
|---|---|---|
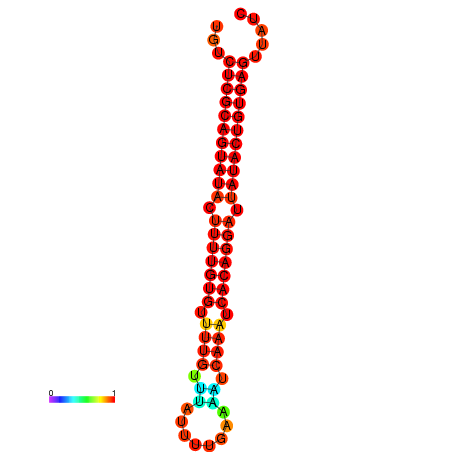 |
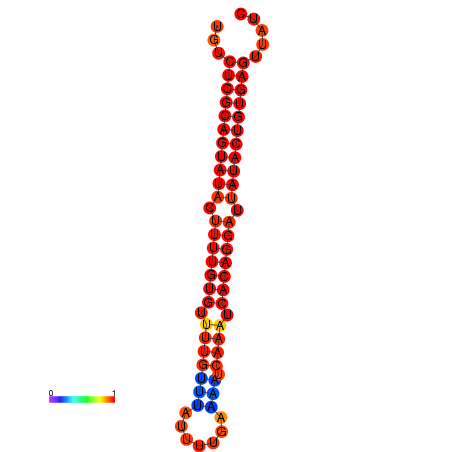 |
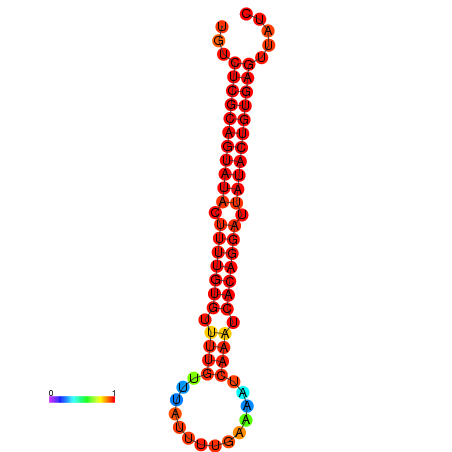 |
| dG=-25.5, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.1, p-value=0.009901 |
|---|---|---|
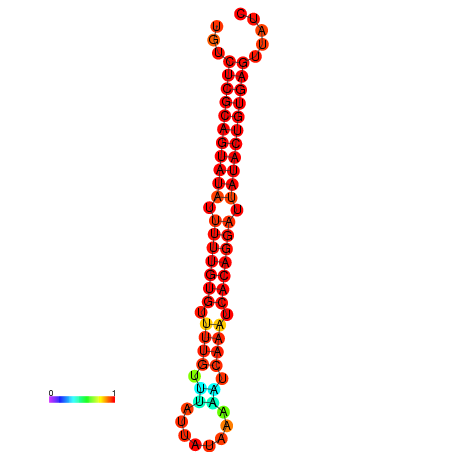 |
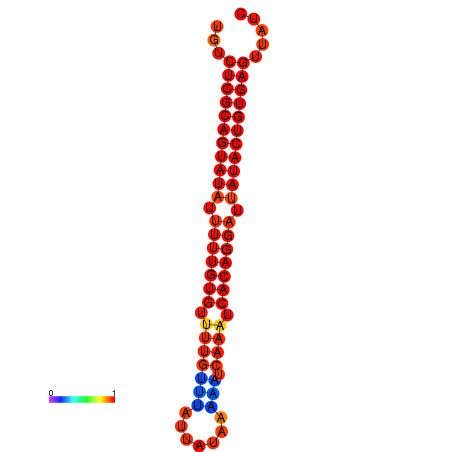 |
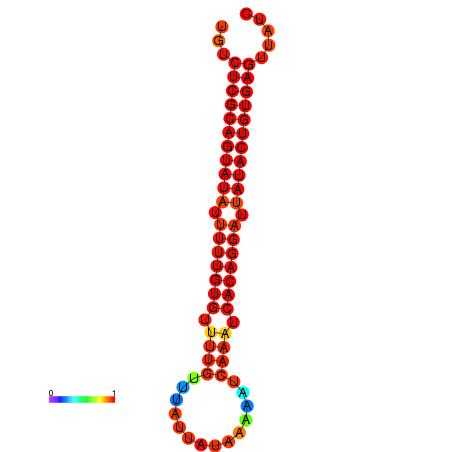 |
| dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-24.6, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
Generated: 03/07/2013 at 05:14 PM