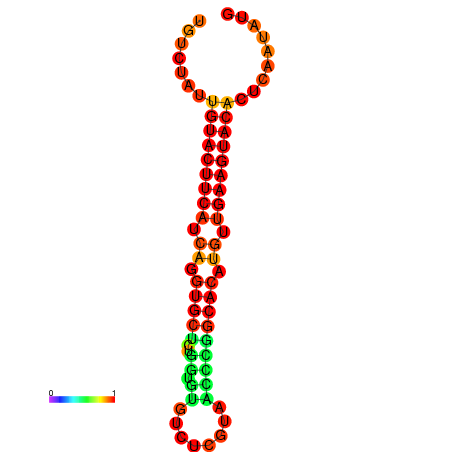
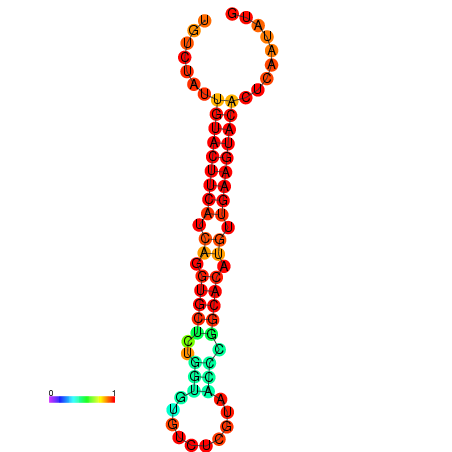
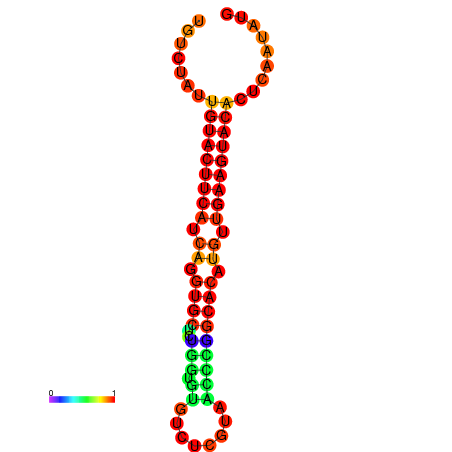
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:7425957-7426059 + | GTAT--------CAAC-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGTC-TCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGATTTGCAT----------------AC |
| droSim1 | chr2L:7217527-7217627 + | GTAT--------CA-C-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGTC-TCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGATTTGCAT----------------AC |
| droSec1 | super_3:2941418-2941520 + | GTAT--------CAAC-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGTC-TCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGATTTGCAT----------------AC |
| droYak2 | chr2L:5473614-5473716 - | GTAT--------CAAC-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGGC-CCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGATTTGCAT----------------AC |
| droEre2 | scaffold_4929:16344683-16344785 + | GTAT--------CAAC-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGGT-CCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGATTTGCAT----------------AC |
| droAna3 | scaffold_12916:15334141-15334251 + | GTATTTTTGAGAAATA-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTCCA-CCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGAATGTCAA----------------AC |
| dp4 | chr4_group1:4175881-4175974 + | G----------------------------------------------------TCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGTC-CTGTAACCCGGCACATGTTGAAGTACACTCAATATGTGGCGAAGTTCGT----------------GC |
| droPer1 | super_5:1086315-1086408 - | G----------------------------------------------------TCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGTC-CTGTAACCCGGCACATGTTGAAGTACACTCAATATGTGGCGAAGTTCGT----------------GC |
| droWil1 | scaffold_180772:5257378-5257513 - | GATT--------CT-CTTTTTTTTTTATTGGCATTTTTAATTTATTGTTGTTGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTCCGTTTGTAACCCGGCACATGTTGAAGTACACTCAATATGTGGCGATCTTCAT------------------ |
| droVir3 | scaffold_12963:15419827-15419927 - | GTTT--------CA-------------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGAC-TTGTAACCCGGCACATGTTGAAGTACACTCAATATGCGTTGATTTGCAT----------------TT |
| droMoj3 | scaffold_6500:18399247-18399346 - | GTGT--------C----------------------------------------TCCCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTCAG-TGGAAACCCGGCACATGTTGAAGTACACTCAATATGCGGTGATTTGCAGTT--------------GC |
| droGri2 | scaffold_15126:5008540-5008648 + | ------------------------------------------------------CTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTCAC-TCGTAACCCGGCACATGTTGAAGTACACTCAATATGCGGTGATTGGCATTTGCTGCGTTTTAATTAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:7425957-7426059 + | GTAT--------CAAC-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGTC-TCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGATTTGCAT----------------AC |
| droSim1 | chr2L:7217527-7217627 + | GTAT--------CA-C-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGTC-TCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGATTTGCAT----------------AC |
| droSec1 | super_3:2941418-2941520 + | GTAT--------CAAC-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGTC-TCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGATTTGCAT----------------AC |
| droYak2 | chr2L:5473614-5473716 - | GTAT--------CAAC-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGGC-CCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGATTTGCAT----------------AC |
| droEre2 | scaffold_4929:16344683-16344785 + | GTAT--------CAAC-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGGT-CCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGATTTGCAT----------------AC |
| droAna3 | scaffold_12916:15334141-15334251 + | GTATTTTTGAGAAATA-----------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTCCA-CCGTAACCCGGCACATGTTGAAGTACACTCAATATGAGGCGAATGTCAA----------------AC |
| dp4 | chr4_group1:4175881-4175974 + | G----------------------------------------------------TCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGTC-CTGTAACCCGGCACATGTTGAAGTACACTCAATATGTGGCGAAGTTCGT----------------GC |
| droPer1 | super_5:1086315-1086408 - | G----------------------------------------------------TCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGTC-CTGTAACCCGGCACATGTTGAAGTACACTCAATATGTGGCGAAGTTCGT----------------GC |
| droWil1 | scaffold_180772:5257378-5257513 - | GATT--------CT-CTTTTTTTTTTATTGGCATTTTTAATTTATTGTTGTTGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTCCGTTTGTAACCCGGCACATGTTGAAGTACACTCAATATGTGGCGATCTTCAT------------------ |
| droVir3 | scaffold_12963:15419827-15419927 - | GTTT--------CA-------------------------------------TGTCTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTGAC-TTGTAACCCGGCACATGTTGAAGTACACTCAATATGCGTTGATTTGCAT----------------TT |
| droMoj3 | scaffold_6500:18399247-18399346 - | GTGT--------C----------------------------------------TCCCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTCAG-TGGAAACCCGGCACATGTTGAAGTACACTCAATATGCGGTGATTTGCAGTT--------------GC |
| droGri2 | scaffold_15126:5008540-5008648 + | ------------------------------------------------------CTCCCATGTCTATTGTACTTCATCAGGTGCTCTGGTGTCAC-TCGTAACCCGGCACATGTTGAAGTACACTCAATATGCGGTGATTGGCATTTGCTGCGTTTTAATTAA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.3, p-value=0.009901 |
|---|---|---|
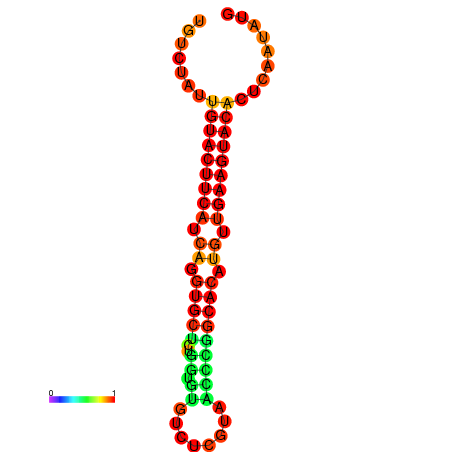 |
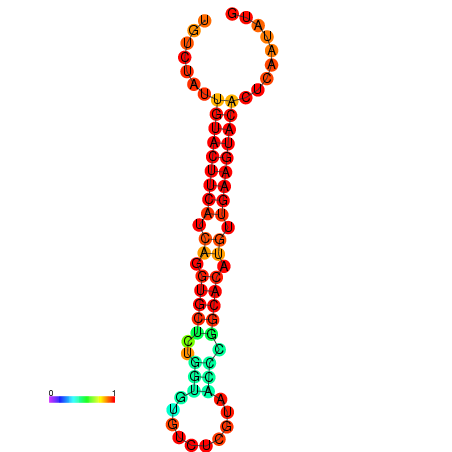 |
 |
| dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.3, p-value=0.009901 |
|---|---|---|
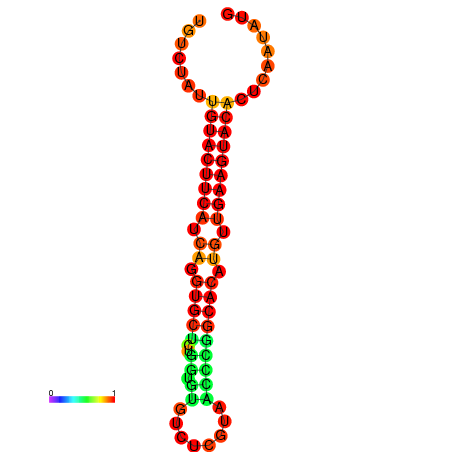 |
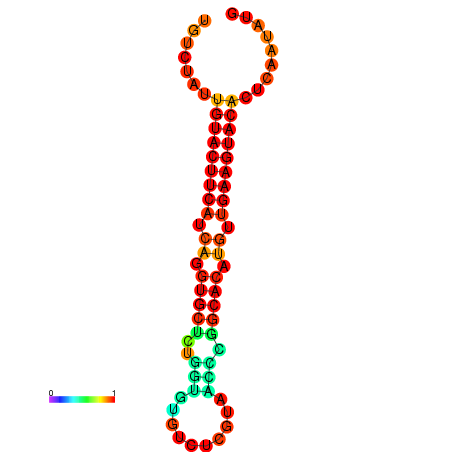 |
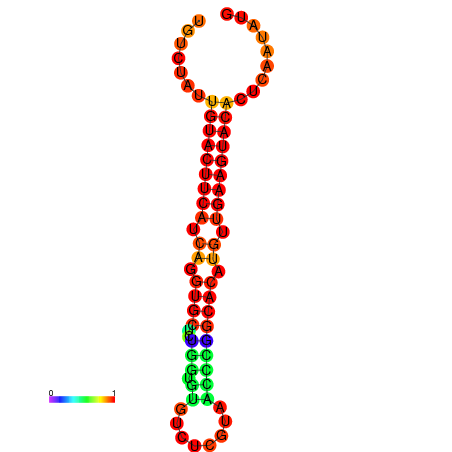 |
| dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.3, p-value=0.009901 |
|---|---|---|
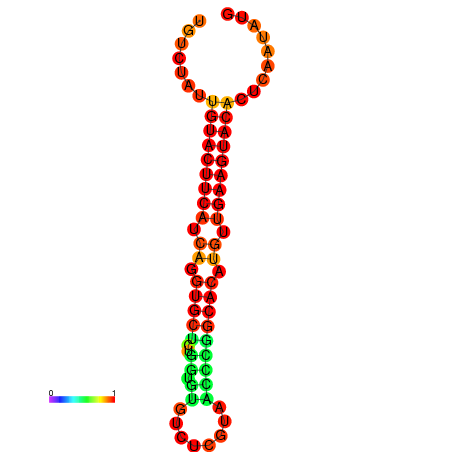 |
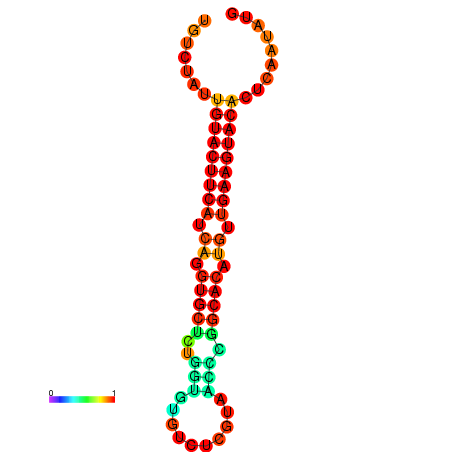 |
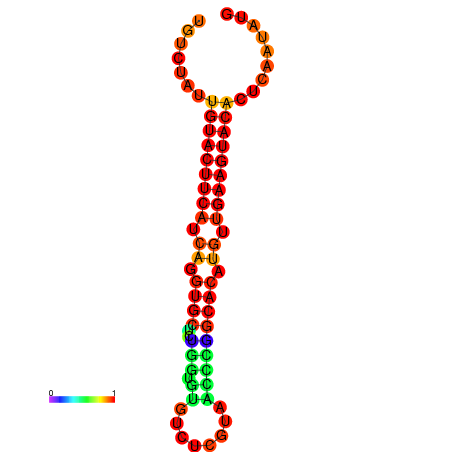 |
| dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.3, p-value=0.009901 |
|---|---|---|
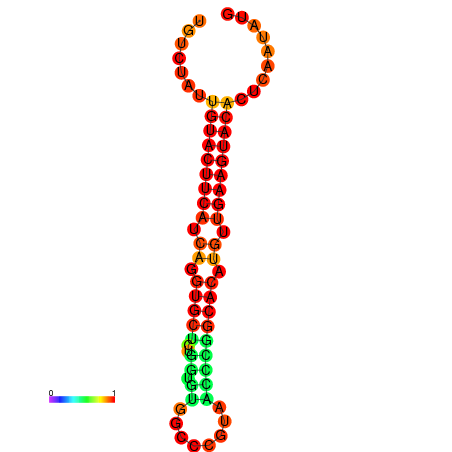 |
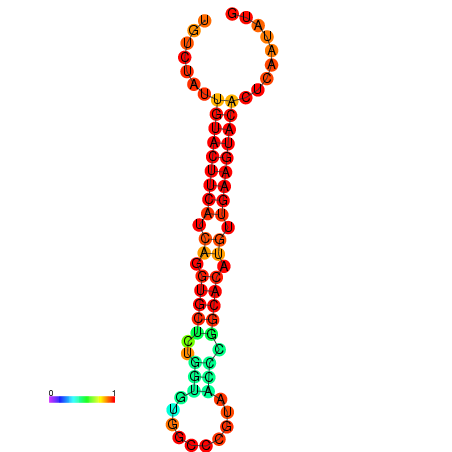 |
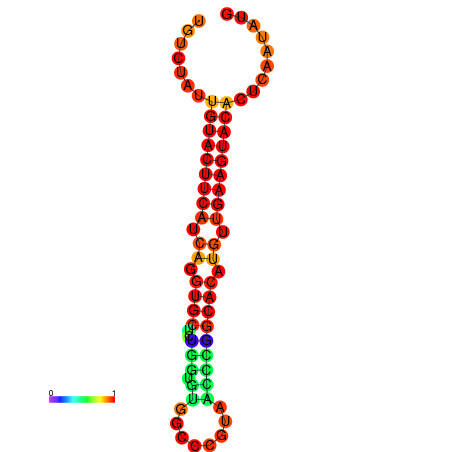 |
| dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.3, p-value=0.009901 |
|---|---|---|
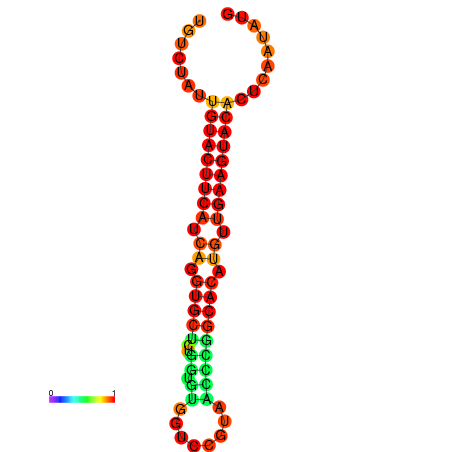 |
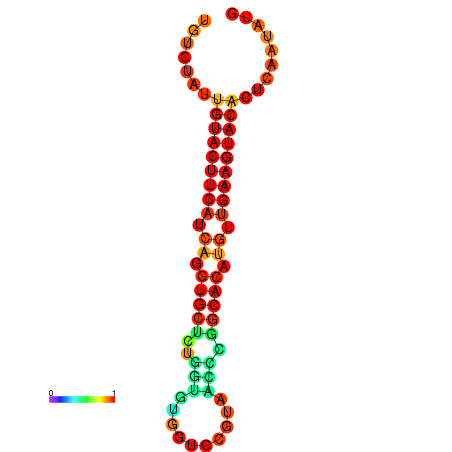 |
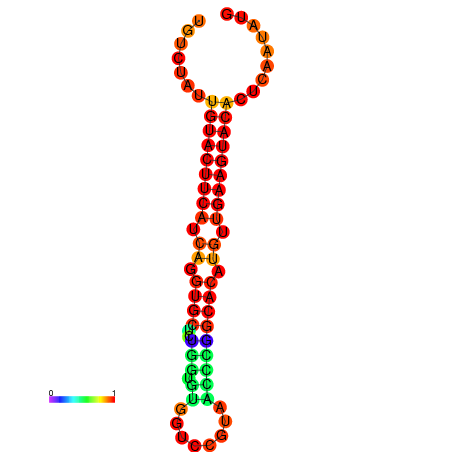 |
| dG=-22.8, p-value=0.009901 |
|---|
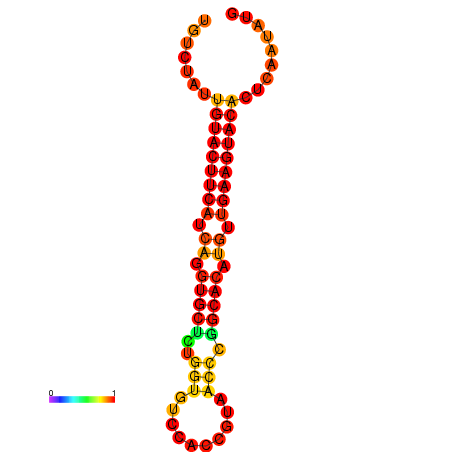 |
| dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.3, p-value=0.009901 |
|---|---|---|
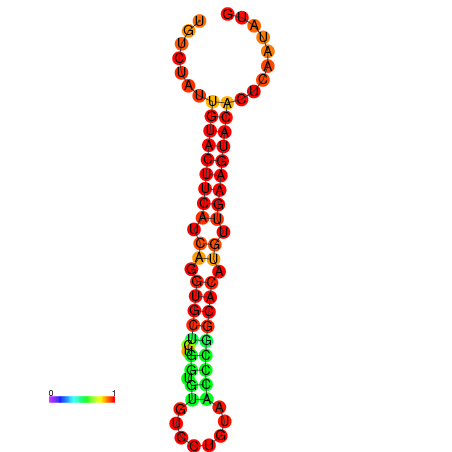 |
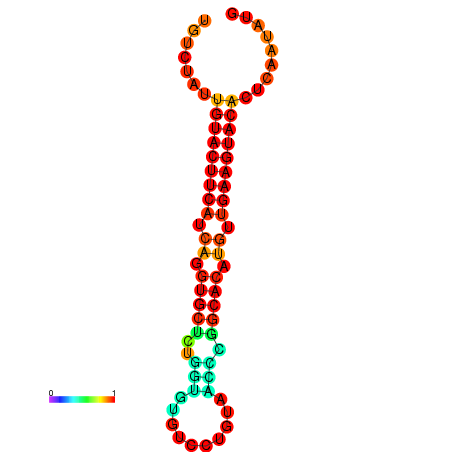 |
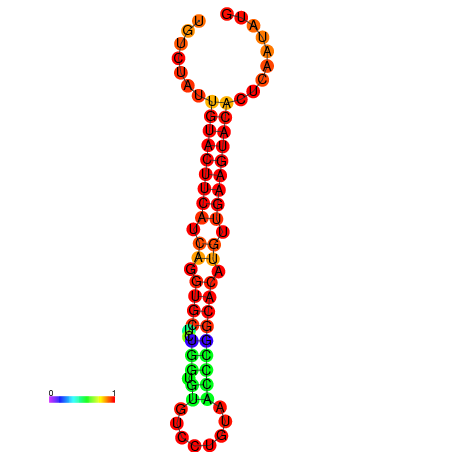 |
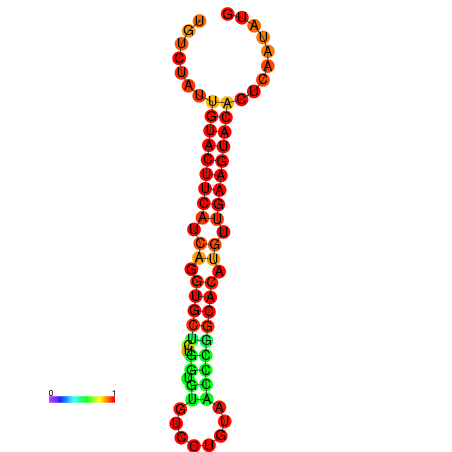
| dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.3, p-value=0.009901 |
|---|---|---|
 |
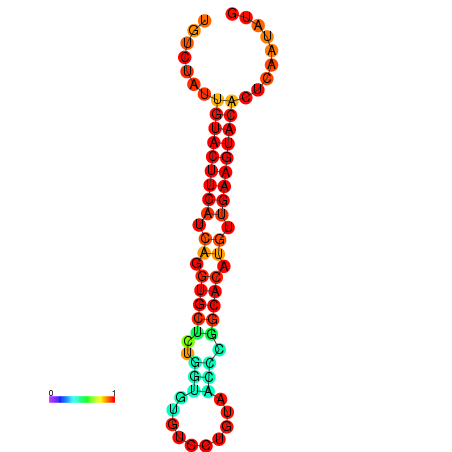 |
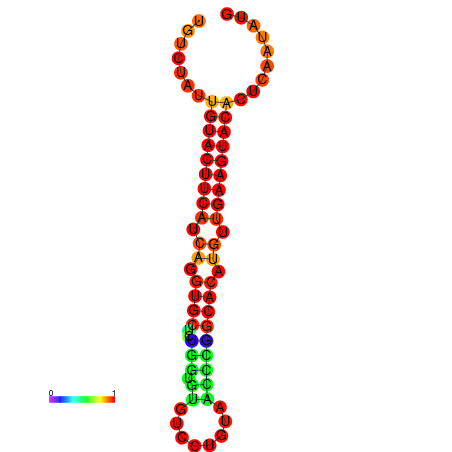 |
| dG=-22.7, p-value=0.009901 |
|---|
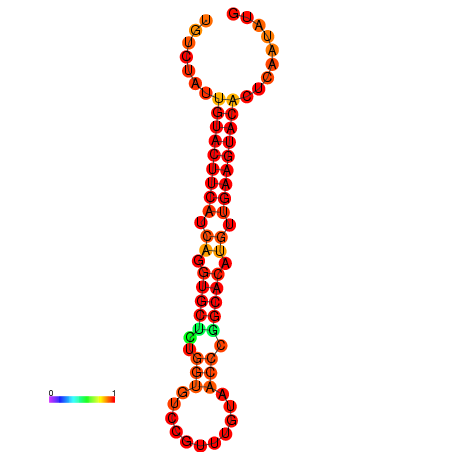 |
| dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.3, p-value=0.009901 |
|---|---|---|
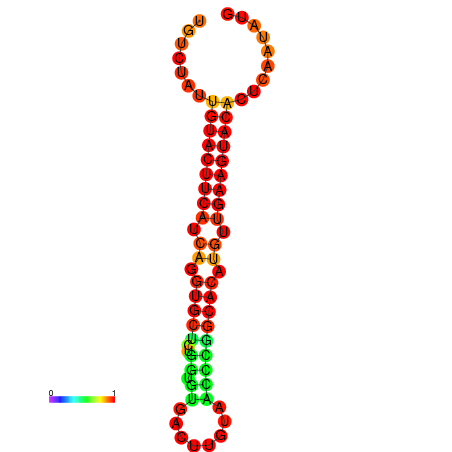 |
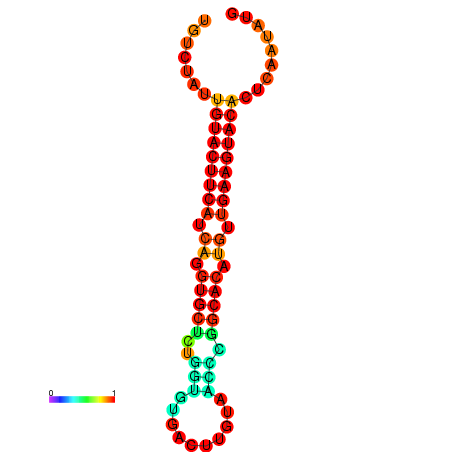 |
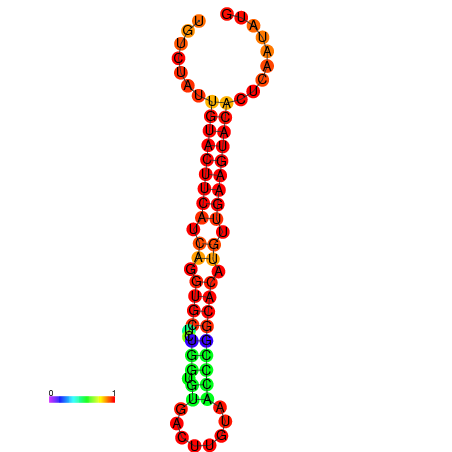 |
| dG=-24.1, p-value=0.009901 |
|---|
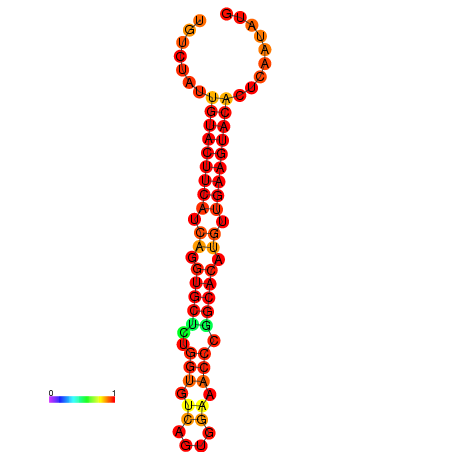 |
| dG=-22.8, p-value=0.009901 |
|---|
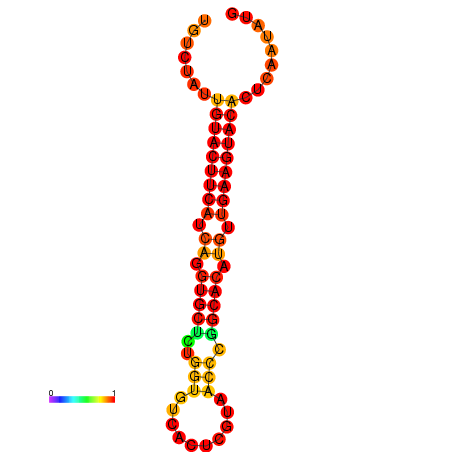 |
Generated: 03/07/2013 at 05:11 PM