| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15549085-15549183 - | GCCA-C------------CTAA-GGTCCCGATCCTGGGATGCATCTTGTGCAGTTATGTTTC--AAT-------CT----CACATCACTGGGCAAAGTGTGTCTCAAGATCCTGGCCACATCG--------------TCG |
| droSim1 | chr2R:14254518-14254616 - | ACCA-C------------CAAA-GGTCCCGATCCTGGGATGCATCTTGTGCAGTTATGTTCC--AAT-------CT----CACATCACTGGGCAAAGTGTGTCTCAAGATCCTGGCCACATCG--------------TCG |
| droSec1 | super_1:13063812-13063910 - | ATCA-C------------CAAA-GGTCCCGATCCTGGGATGCATCTTGTGCAGTTATGTTTC--AAT-------CT----CACATCACTGGGCAAAGTGTGTCTCAAGATCCTGGCCACATCG--------------TCG |
| droYak2 | chr2R:13623606-13623704 + | ACCA-T------------CAAA-GGTCCTGATCCTGGGATGCATCTTGTGCAGTCATGTTTC--CAT-------CT----CACATCACTGGGCAAAGTGTGTCTCAAGATCTTGGCCACTTCG--------------ACG |
| droEre2 | scaffold_4845:9749329-9749427 - | ACCC-A------------CCAA-GGTCCTGATCCTGGGATGCATCTTGTGCAGTTATGTTTC--TAT-------CT----CACATCACTGGGCAAAGTGTGTCTCAAGATCCTGGCCACTTCG--------------ACG |
| droAna3 | scaffold_13266:9731210-9731313 + | GCCC-C------------CAACTGGCTACGATCTTGGGACGCATTTTGTGCAGTTATGCACC--AAA-AATA------TATCCATCACTGGGCAAAGTGTGTCTCAGGATCTTGGCCATCTCA--------------ACG |
| dp4 | chr3:12677741-12677846 - | ACGA-CGA--------TACCGA-GGCAACGATCCTGGGATGCATTTTGTGCAGTTATGTCTA--CGTGATCA------TCCTCATCACTGGGCAAAGTGTGTCTCAGGATTGCAGCCCCGCCA--------------A-- |
| droPer1 | super_2:8905820-8905925 + | ACGA-CGA--------TACCGA-GGCAACGATCCTGGGATGCATTTTGTGCAGTTATGTCTA--CGTGATCA------TCCTCATCACTGGGCAAAGTGTGTCTCAGGATTGCAGCCCCGCCA--------------A-- |
| droWil1 | scaffold_180701:1651920-1652025 - | TCGA-A------------TTTA-GACCATGATTTTGGGATGCATTTTGTGCAGTTATGTCTTCATGTGATAATTCT----CTCATCACTGGGCAAAGTGTGTCTCATAATTTTGGTAACCATA--------------T-- |
| droVir3 | scaffold_12875:4111929-4112038 + | AATAAAAATACCAGAATCTCAT-AGCTCTGCTTTTGGGATGCATTTTGTGCGGTTATGTCTG--CCT-------CCAAAACCCATCACTGGGCAAAGTGTGTCTCAAAATTGTGGCTACC-------------------- |
| droMoj3 | scaffold_6496:20936961-20937077 - | AACT-C------------TCGC-AGCTCTACTTTTGGGATGCATTTTGTGCAGATGTCTAAC--GAT-------TGGAAACCCATCACTGGGCAAAGTGTGTCTCAAAATTGGGGCAACTAGCACAAAGGAGGAGTGTTT |
| droGri2 | scaffold_15112:2109003-2109099 - | AATT-T------------TCAT-AGCTCTGCTTTTGGGATGCATTCTGTACAGTAATGTTGA--CTCTTGTA------AAACCATCACTGGGCAAAGTGTGTCTCAAAATTGTGGCAA---------------------G |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15549085-15549183 - | GCCA-C------------CTAA-GGTCCCGATCCTGGGATGCATCTTGTGCAGTTATGTTTC--AAT-------CT----CACATCACTGGGCAAAGTGTGTCTCAAGATCCTGGCCACATCG--------------TCG |
| droSim1 | chr2R:14254518-14254616 - | ACCA-C------------CAAA-GGTCCCGATCCTGGGATGCATCTTGTGCAGTTATGTTCC--AAT-------CT----CACATCACTGGGCAAAGTGTGTCTCAAGATCCTGGCCACATCG--------------TCG |
| droSec1 | super_1:13063812-13063910 - | ATCA-C------------CAAA-GGTCCCGATCCTGGGATGCATCTTGTGCAGTTATGTTTC--AAT-------CT----CACATCACTGGGCAAAGTGTGTCTCAAGATCCTGGCCACATCG--------------TCG |
| droYak2 | chr2R:13623606-13623704 + | ACCA-T------------CAAA-GGTCCTGATCCTGGGATGCATCTTGTGCAGTCATGTTTC--CAT-------CT----CACATCACTGGGCAAAGTGTGTCTCAAGATCTTGGCCACTTCG--------------ACG |
| droEre2 | scaffold_4845:9749329-9749427 - | ACCC-A------------CCAA-GGTCCTGATCCTGGGATGCATCTTGTGCAGTTATGTTTC--TAT-------CT----CACATCACTGGGCAAAGTGTGTCTCAAGATCCTGGCCACTTCG--------------ACG |
| droAna3 | scaffold_13266:9731210-9731313 + | GCCC-C------------CAACTGGCTACGATCTTGGGACGCATTTTGTGCAGTTATGCACC--AAA-AATA------TATCCATCACTGGGCAAAGTGTGTCTCAGGATCTTGGCCATCTCA--------------ACG |
| dp4 | chr3:12677741-12677846 - | ACGA-CGA--------TACCGA-GGCAACGATCCTGGGATGCATTTTGTGCAGTTATGTCTA--CGTGATCA------TCCTCATCACTGGGCAAAGTGTGTCTCAGGATTGCAGCCCCGCCA--------------A-- |
| droPer1 | super_2:8905820-8905925 + | ACGA-CGA--------TACCGA-GGCAACGATCCTGGGATGCATTTTGTGCAGTTATGTCTA--CGTGATCA------TCCTCATCACTGGGCAAAGTGTGTCTCAGGATTGCAGCCCCGCCA--------------A-- |
| droWil1 | scaffold_180701:1651920-1652025 - | TCGA-A------------TTTA-GACCATGATTTTGGGATGCATTTTGTGCAGTTATGTCTTCATGTGATAATTCT----CTCATCACTGGGCAAAGTGTGTCTCATAATTTTGGTAACCATA--------------T-- |
| droVir3 | scaffold_12875:4111929-4112038 + | AATAAAAATACCAGAATCTCAT-AGCTCTGCTTTTGGGATGCATTTTGTGCGGTTATGTCTG--CCT-------CCAAAACCCATCACTGGGCAAAGTGTGTCTCAAAATTGTGGCTACC-------------------- |
| droMoj3 | scaffold_6496:20936961-20937077 - | AACT-C------------TCGC-AGCTCTACTTTTGGGATGCATTTTGTGCAGATGTCTAAC--GAT-------TGGAAACCCATCACTGGGCAAAGTGTGTCTCAAAATTGGGGCAACTAGCACAAAGGAGGAGTGTTT |
| droGri2 | scaffold_15112:2109003-2109099 - | AATT-T------------TCAT-AGCTCTGCTTTTGGGATGCATTCTGTACAGTAATGTTGA--CTCTTGTA------AAACCATCACTGGGCAAAGTGTGTCTCAAAATTGTGGCAA---------------------G |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
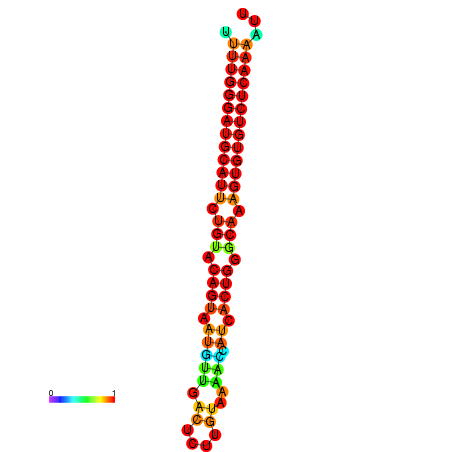
| dG=-24.6, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|
 |
 |
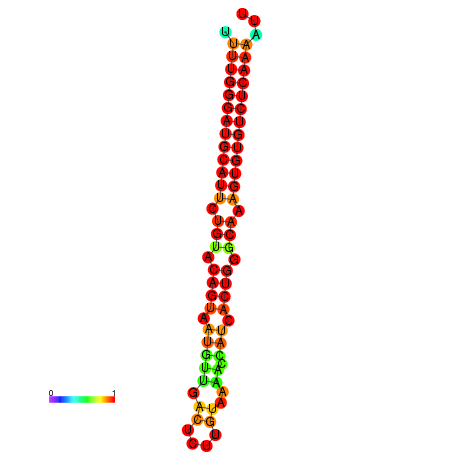
| dG=-24.6, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|
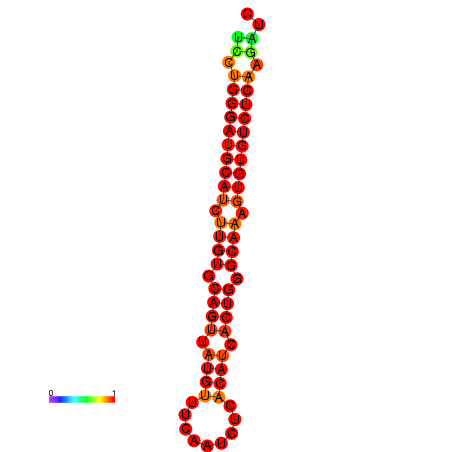
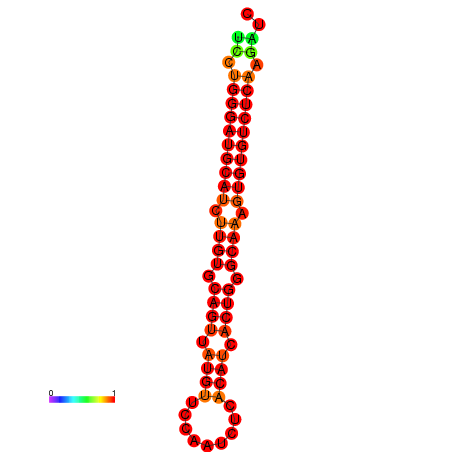 |
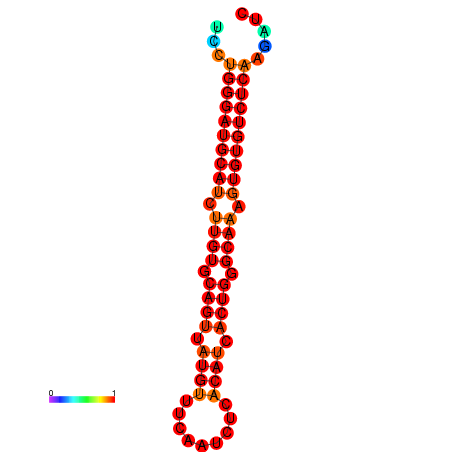
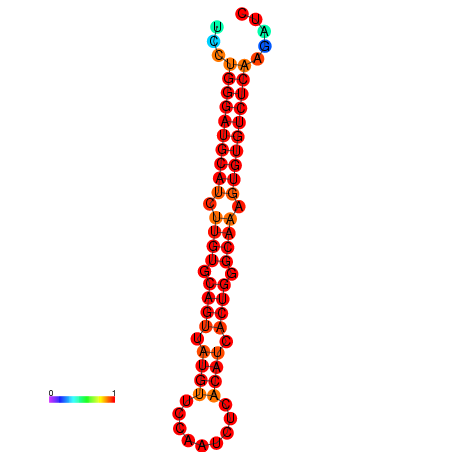 |
| dG=-24.6, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|
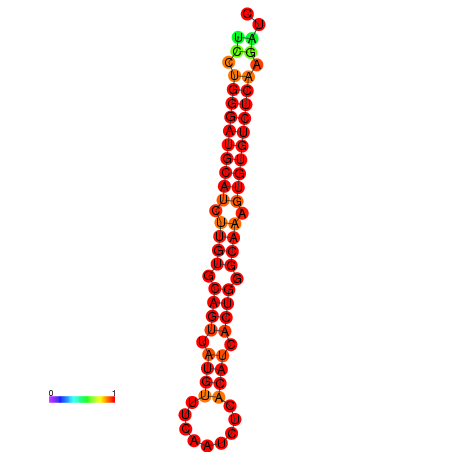 |
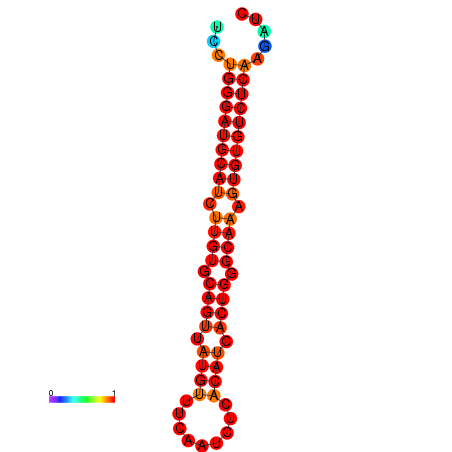 |
| dG=-24.6, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|
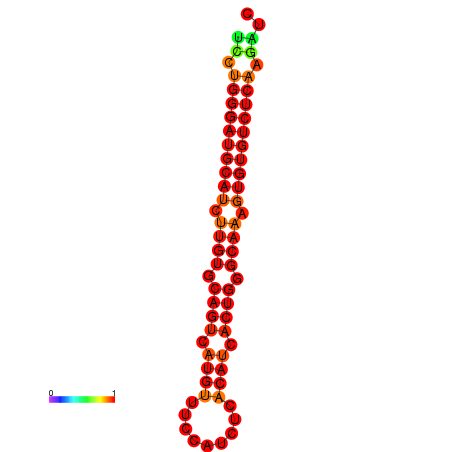 |
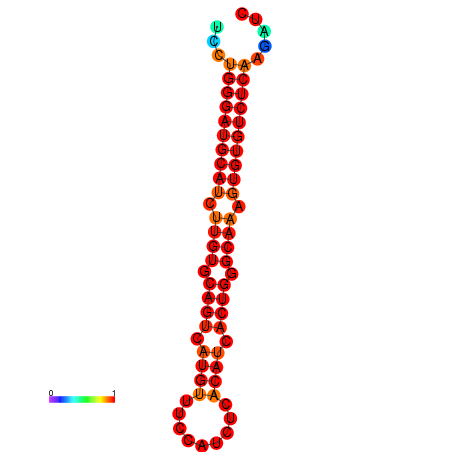 |
| dG=-24.6, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|
 |
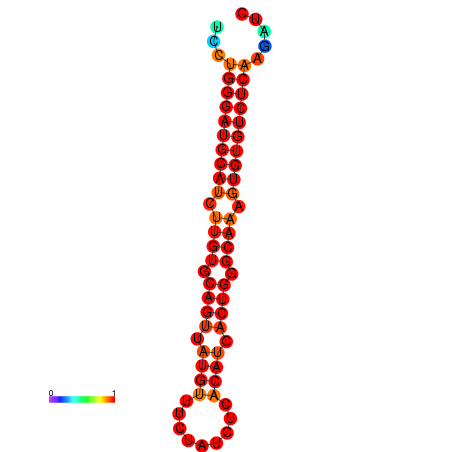 |
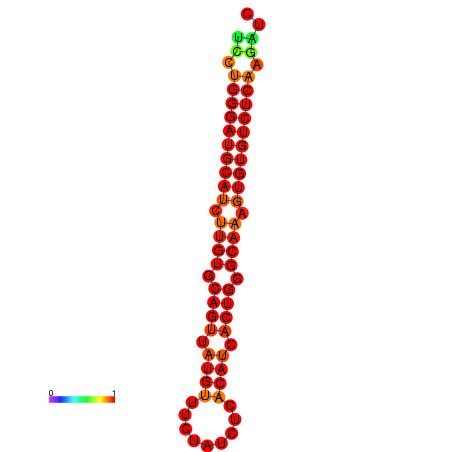
| dG=-32.3, p-value=0.009901 | dG=-31.3, p-value=0.009901 |
|---|---|
 |
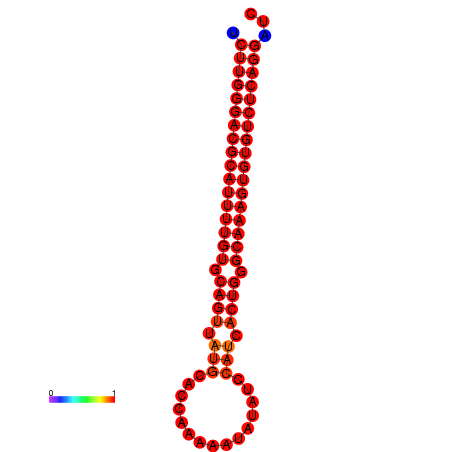 |
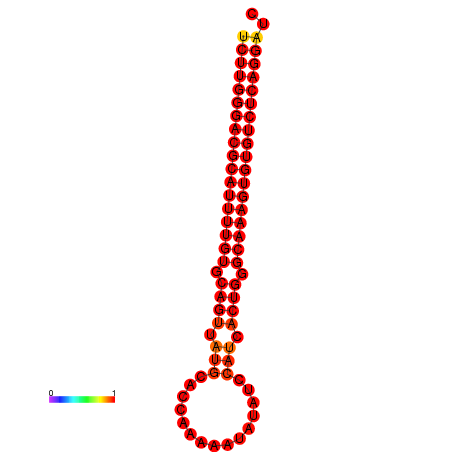
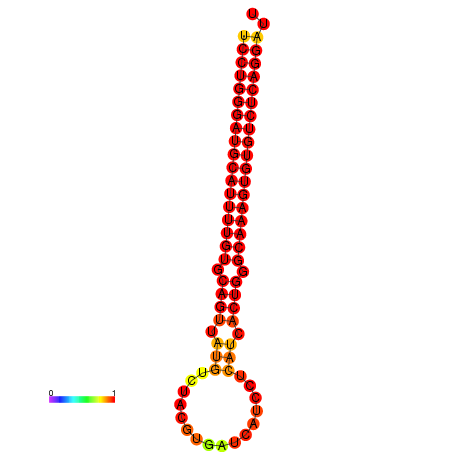
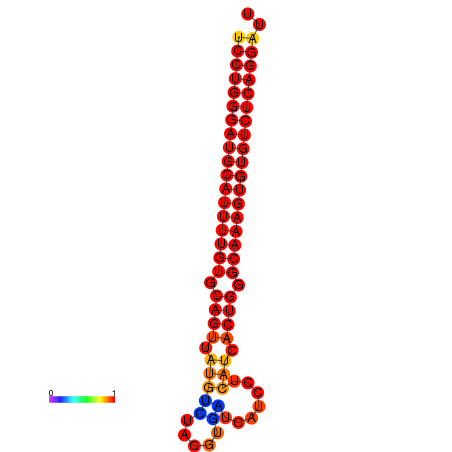
| dG=-33.3, p-value=0.009901 | dG=-32.4, p-value=0.009901 | dG=-32.3, p-value=0.009901 |
|---|---|---|
 |
 |
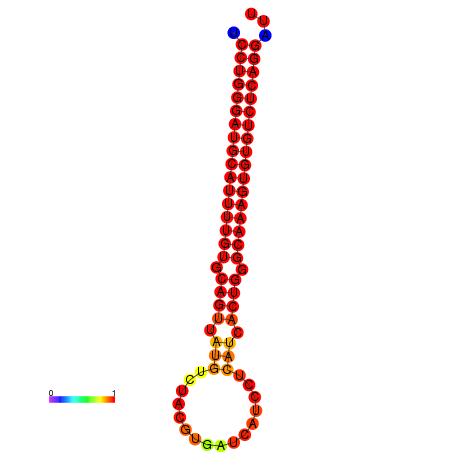 |
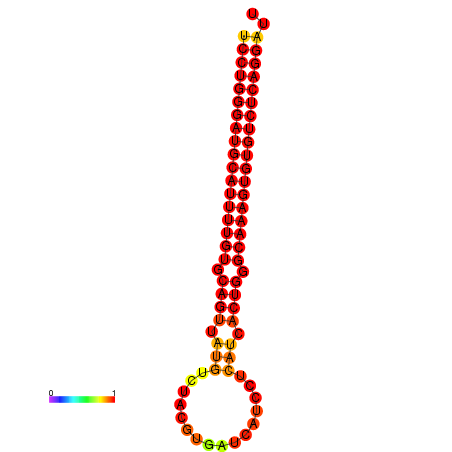
| dG=-33.3, p-value=0.009901 | dG=-32.4, p-value=0.009901 | dG=-32.3, p-value=0.009901 |
|---|---|---|
 |
 |
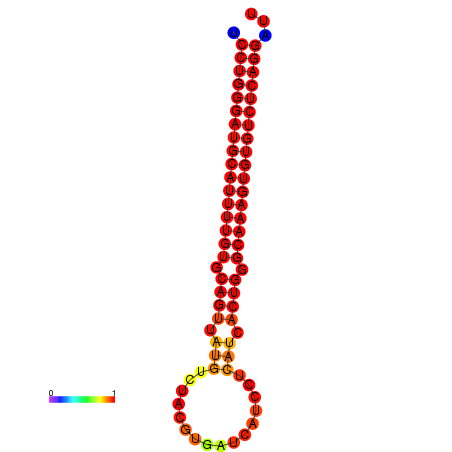 |
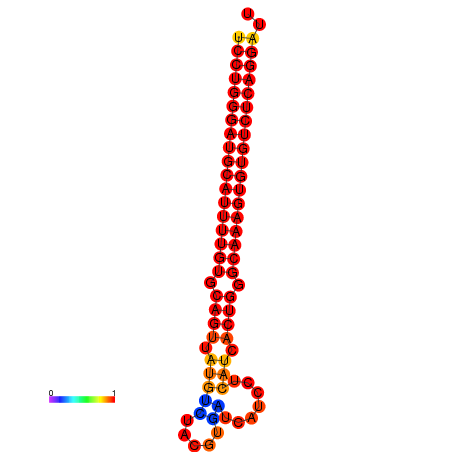
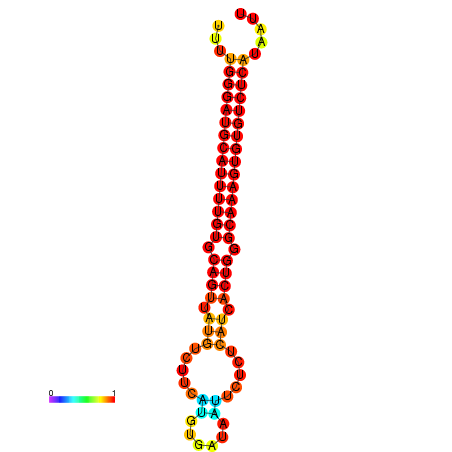
| dG=-25.9, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|---|---|---|
 |
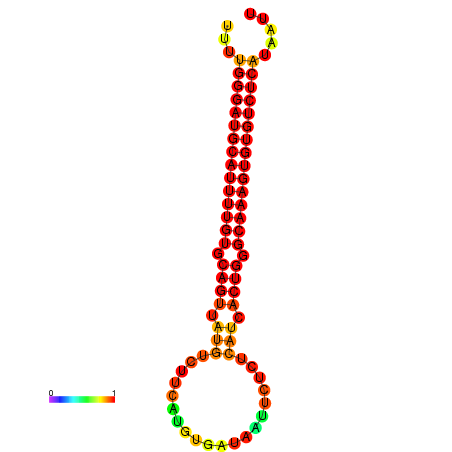 |
 |
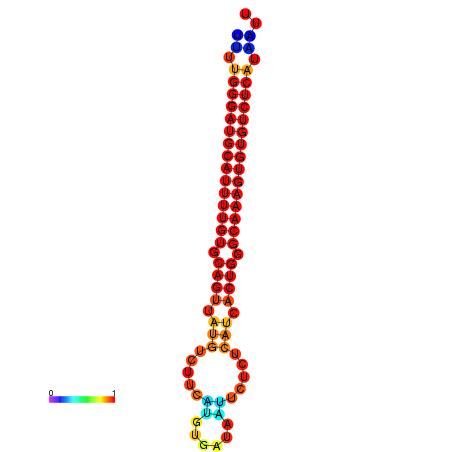 |
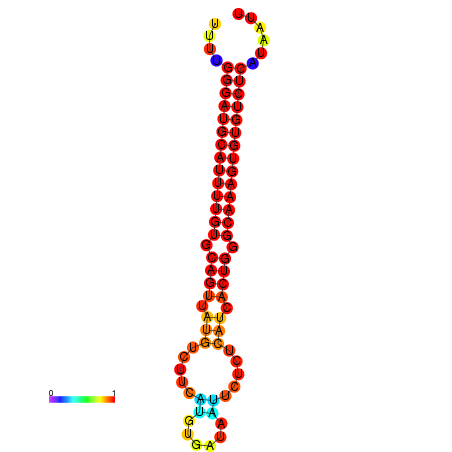 |
| dG=-27.0, p-value=0.009901 | dG=-26.7, p-value=0.009901 |
|---|---|
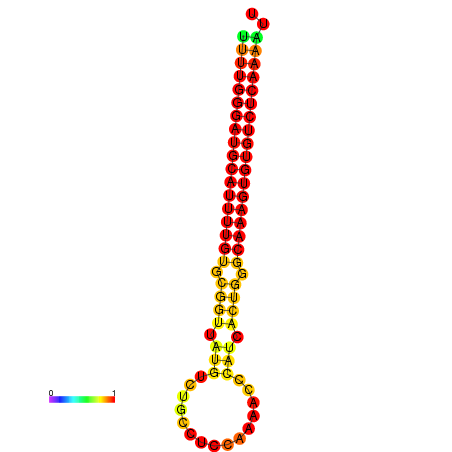 |
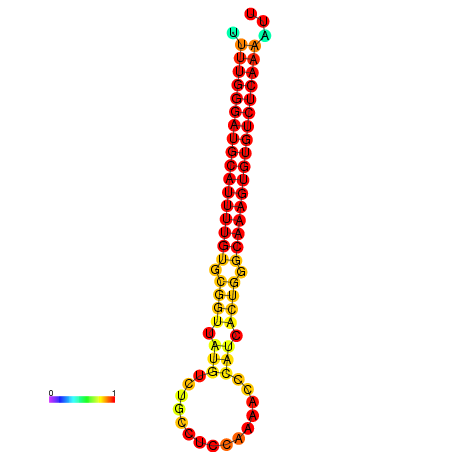 |
| dG=-26.7, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
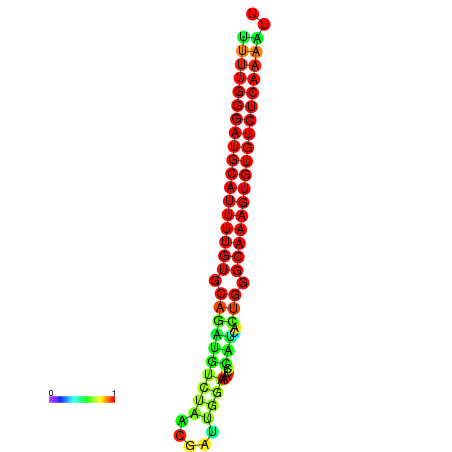 |
| dG=-22.7, p-value=0.009901 | dG=-22.7, p-value=0.009901 | dG=-22.6, p-value=0.009901 | dG=-22.4, p-value=0.009901 | dG=-22.4, p-value=0.009901 |
|---|---|---|---|---|
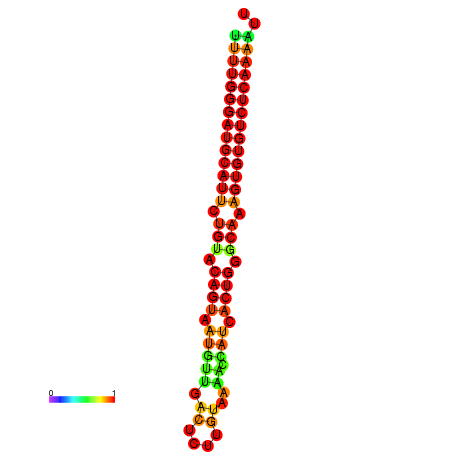 |
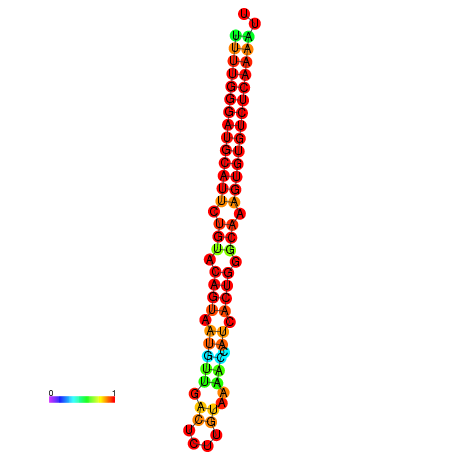 |
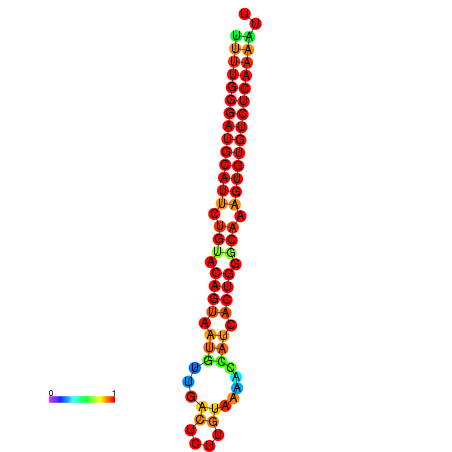 |
 |
 |
Generated: 03/07/2013 at 05:10 PM