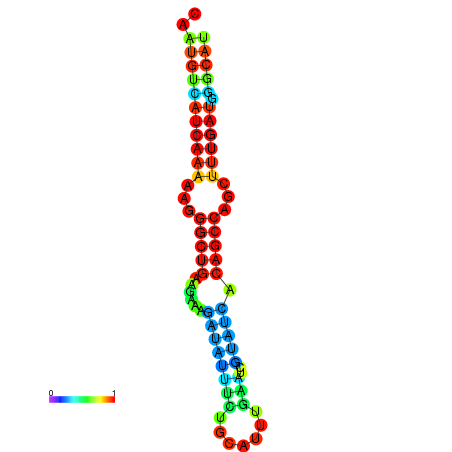
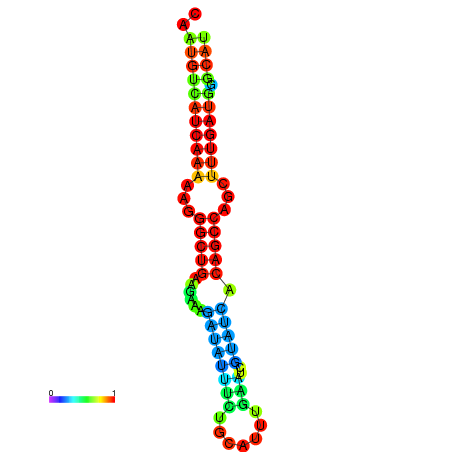
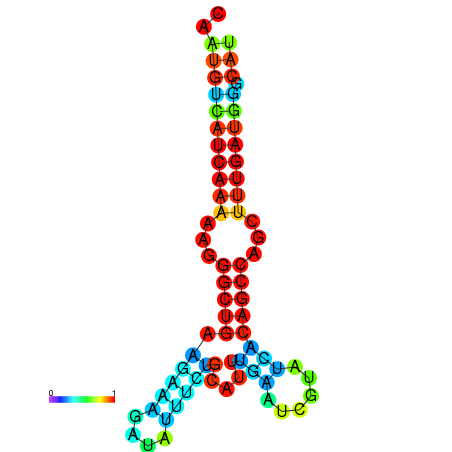
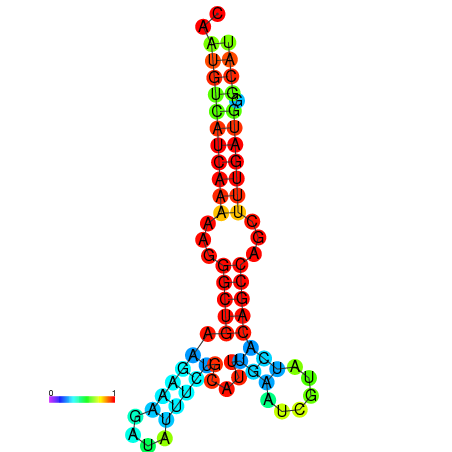
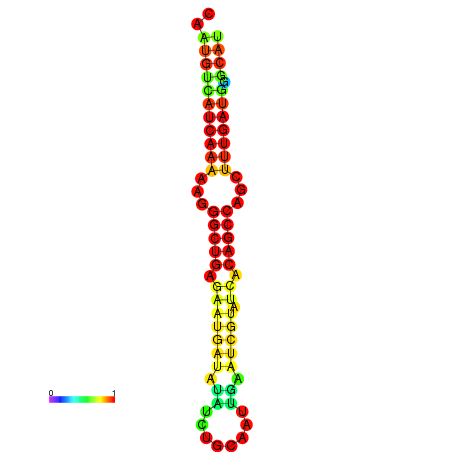
| TGCGTGCGTGCTTGCGTATCGG---CACTTGGTATCCTTACTTTCAGTGTCATCAAAAATGGCTGAAGAAAGATATATTTGCATTTGAAGCGTATCACAGCCAGCTTTGATGGGCATTGCAATGAGCATCGATTGCA----GTTTCG | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ..............................................................................................TCACAGCCAGCTTTGATGGGCA............................... | 22 | 1 | 1141.00 | 1141 | 1035 | 106 |
| ............................................................................................TATCACAGCCAGCTTTGATGGG................................. | 22 | 1 | 48.00 | 48 | 34 | 14 |
| ..............................................................................................TCACAGCCAGCTTTGATGGGC................................ | 21 | 1 | 40.00 | 40 | 28 | 12 |
| ............................................................................................TATCACAGCCAGCTTTGATGGGC................................ | 23 | 1 | 39.00 | 39 | 33 | 6 |
| ..............................................................................................TCACAGCCAGCTTTGATGGG................................. | 20 | 1 | 26.00 | 26 | 16 | 10 |
| ............................................................................................TATCACAGCCAGCTTTGATGGGCA............................... | 24 | 1 | 22.00 | 22 | 18 | 4 |
| ................................................................................................ACAGCCAGCTTTGATGGGCA............................... | 20 | 1 | 16.00 | 16 | 15 | 1 |
| ..............................................................................................TCACAGCCAGCTTTGATGGGCAT.............................. | 23 | 1 | 9.00 | 9 | 6 | 3 |
| .................................................TCATCAAAAATGGCTGAAG............................................................................... | 19 | 1 | 5.00 | 5 | 0 | 5 |
| ............................................................................................TATCACAGCCAGCTTTGA..................................... | 18 | 5 | 4.60 | 23 | 23 | 0 |
| ..............................................................................................TCACAGCCAGCTTTGATGG.................................. | 19 | 1 | 4.00 | 4 | 3 | 1 |
| .............................................................................................ATCACAGCCAGCTTTGATGGGC................................ | 22 | 1 | 4.00 | 4 | 2 | 2 |
| .................................................TCATCAAAAATGGCTGAAGA.............................................................................. | 20 | 1 | 3.00 | 3 | 0 | 3 |
| .................................................TCATCAAAAATGGCTGAAGAAAG........................................................................... | 23 | 1 | 3.00 | 3 | 0 | 3 |
| .................................................TCATCAAAAATGGCTGAAGAAAGATA........................................................................ | 26 | 1 | 3.00 | 3 | 3 | 0 |
| ............................................................................................TATCACAGCCAGCTTTGATGG.................................. | 21 | 1 | 3.00 | 3 | 3 | 0 |
| ...............................................................................................CACAGCCAGCTTTGATGGGCA............................... | 21 | 1 | 3.00 | 3 | 3 | 0 |
| .................................................TCATCAAAAATGGCTGAAGAAAGA.......................................................................... | 24 | 1 | 3.00 | 3 | 3 | 0 |
| .................................................................................................CAGCCAGCTTTGATGGGCA............................... | 19 | 1 | 2.00 | 2 | 2 | 0 |
| .................................................TCATCAAAAATGGCTGAAGAAAGAT......................................................................... | 25 | 1 | 2.00 | 2 | 2 | 0 |
| ..................................................................................................AGCCAGCTTTGATGGGCA............................... | 18 | 1 | 2.00 | 2 | 2 | 0 |
| ...............................................................................................CACAGCCAGCTTTGATGGGCAT.............................. | 22 | 1 | 2.00 | 2 | 2 | 0 |
| .................................................TCATCAAAAATGGCTGAAGAAA............................................................................ | 22 | 1 | 2.00 | 2 | 0 | 2 |
| .............................................................................................ATCACAGCCAGCTTTGATGGGCA............................... | 23 | 1 | 2.00 | 2 | 2 | 0 |
| ................................................................................................ACAGCCAGCTTTGATGGGC................................ | 19 | 1 | 1.00 | 1 | 1 | 0 |
| ...........................................................................TATTTGCATTTGAAGCGTA..................................................... | 19 | 1 | 1.00 | 1 | 1 | 0 |
| ................................................................................................ACAGCCAGCTTTGATGGG................................. | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ..................................................CATCAAAAATGGCTGAAGAAAGATA........................................................................ | 25 | 1 | 1.00 | 1 | 1 | 0 |