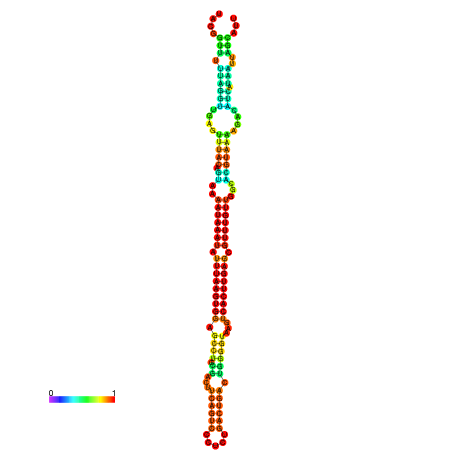
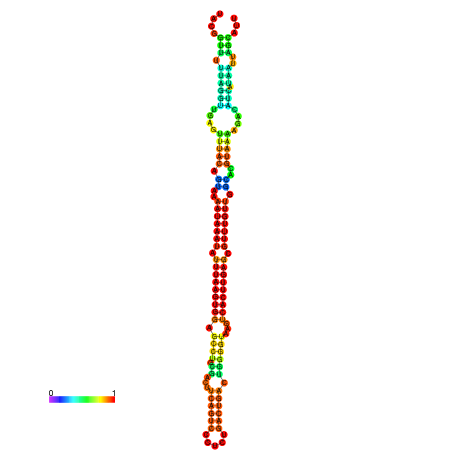
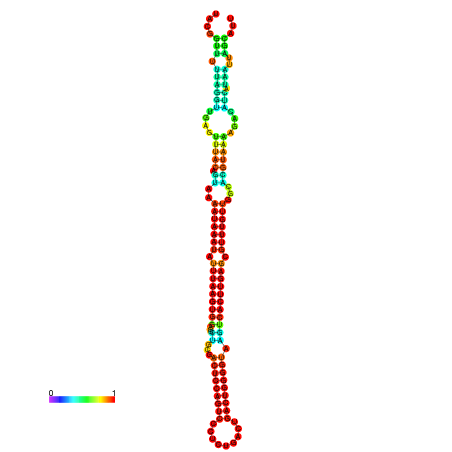
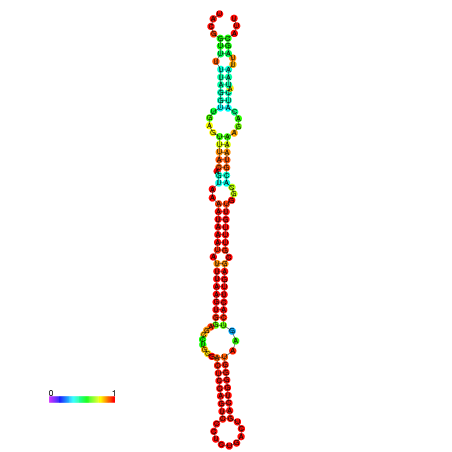
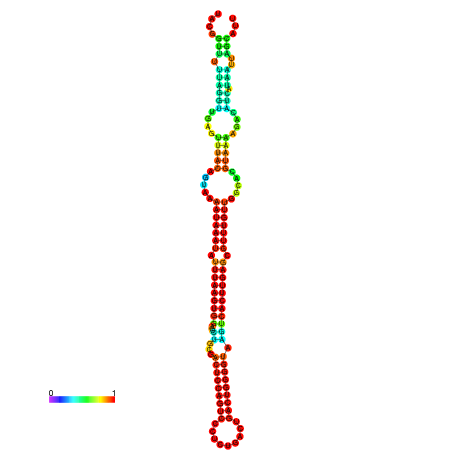
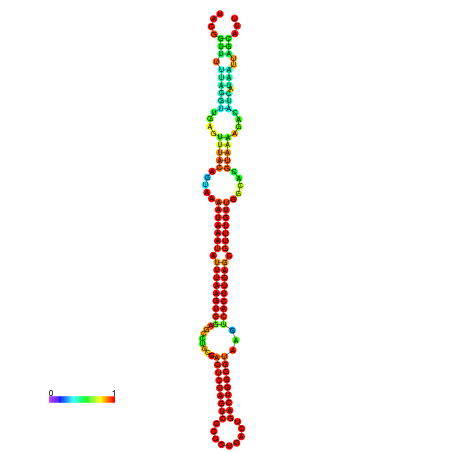
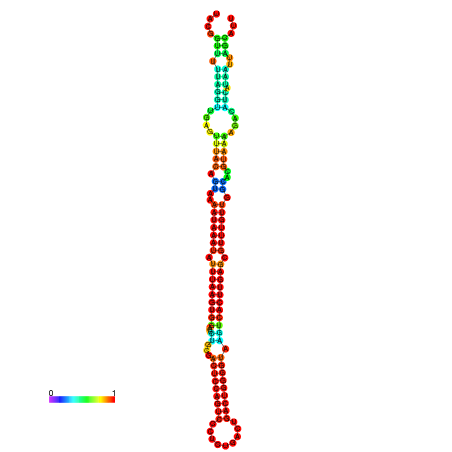
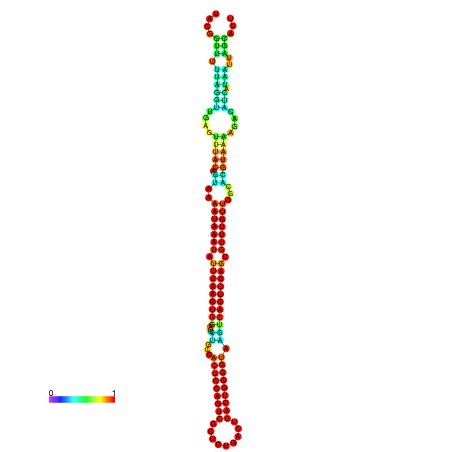
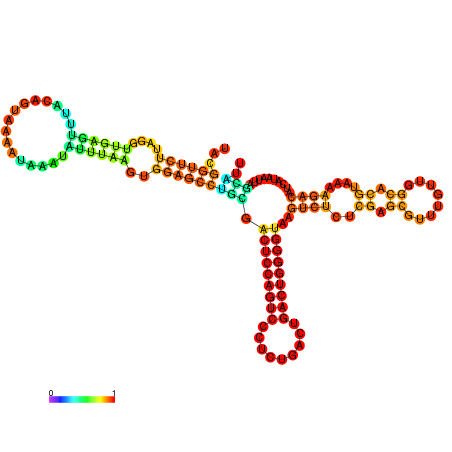
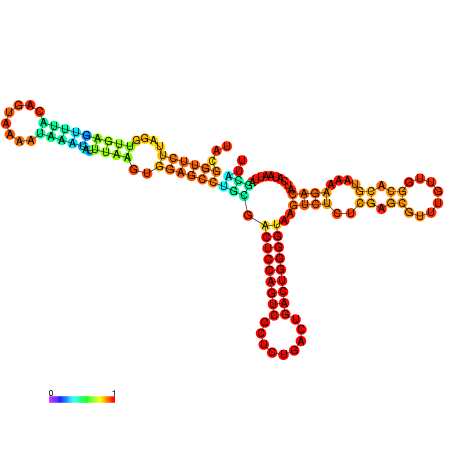
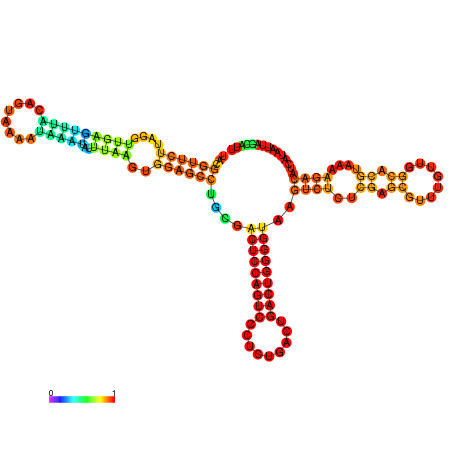
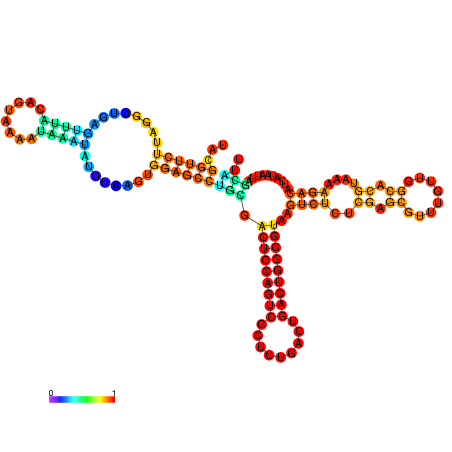
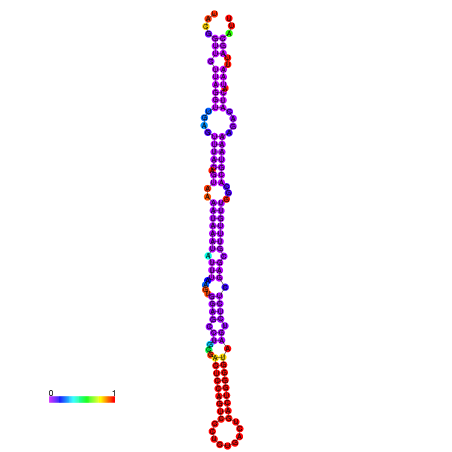
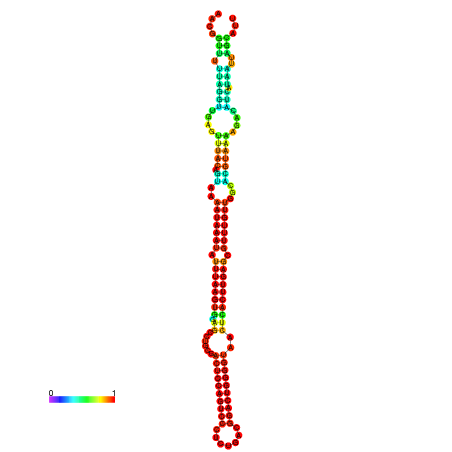
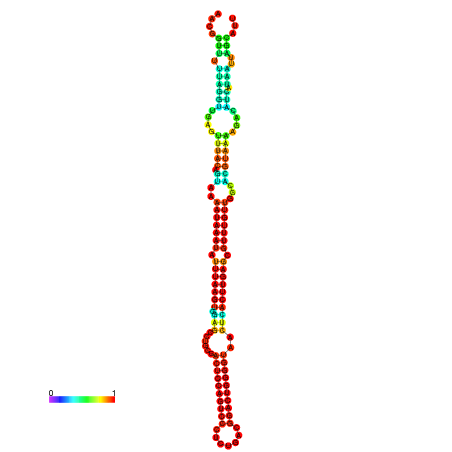
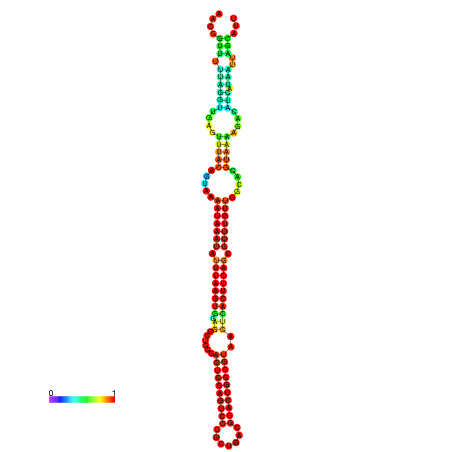
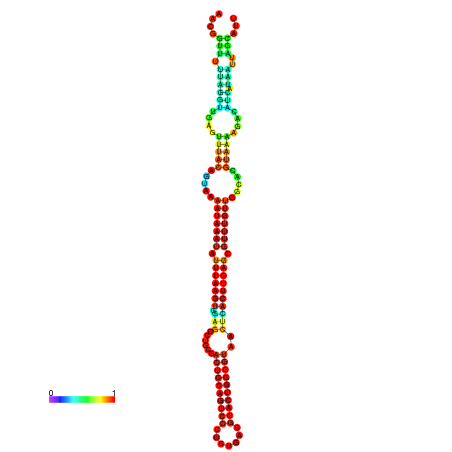
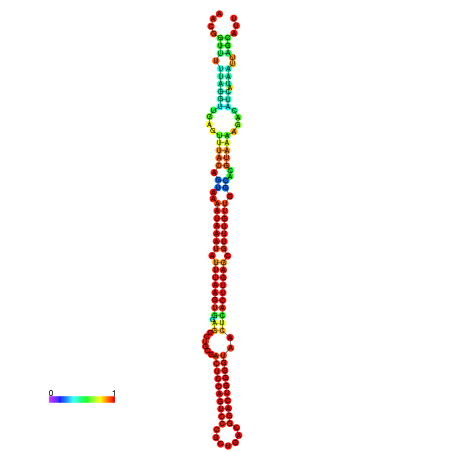
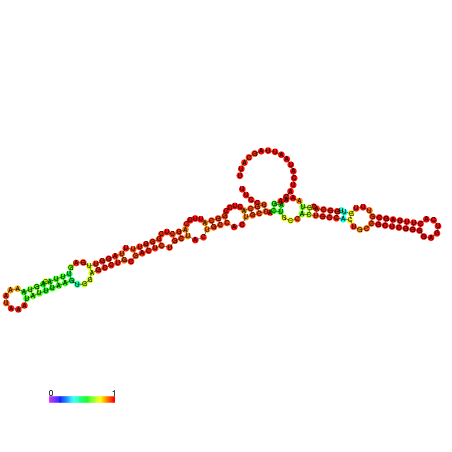
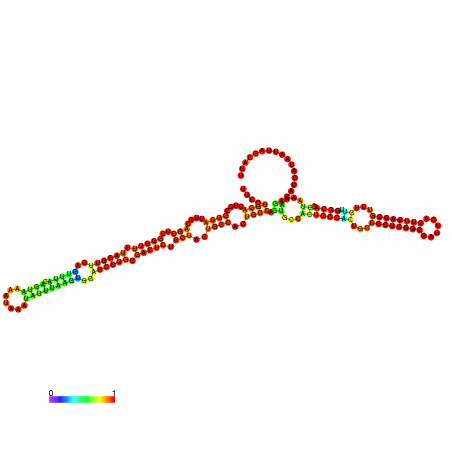
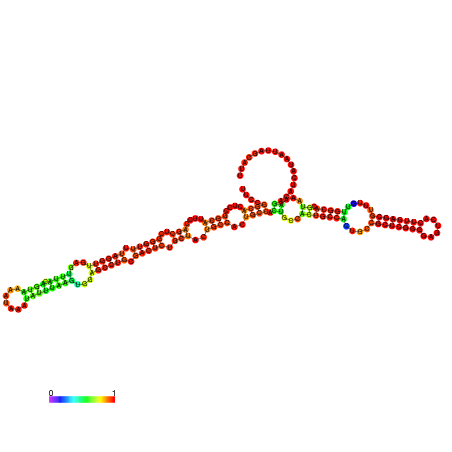
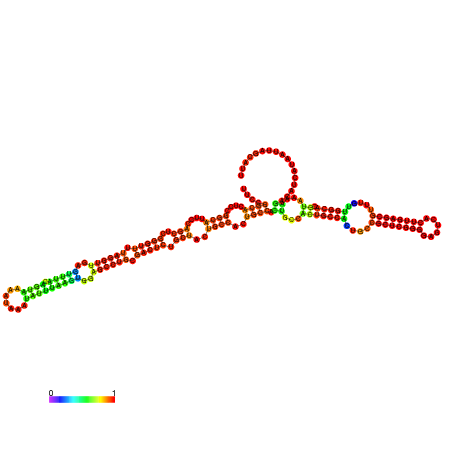
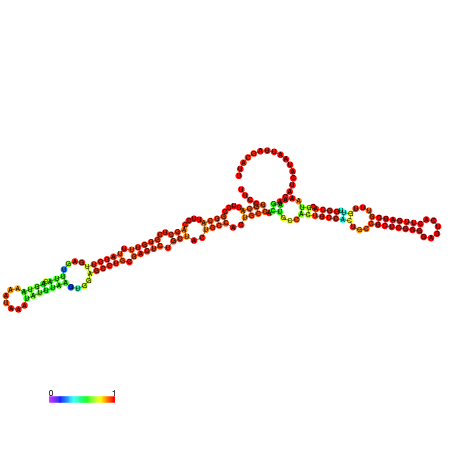
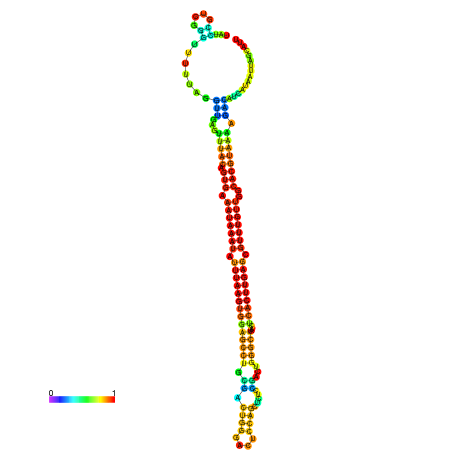
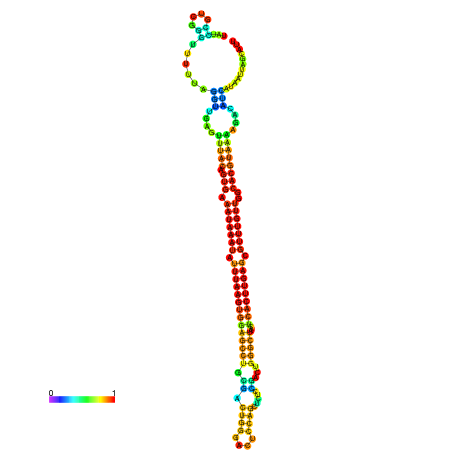
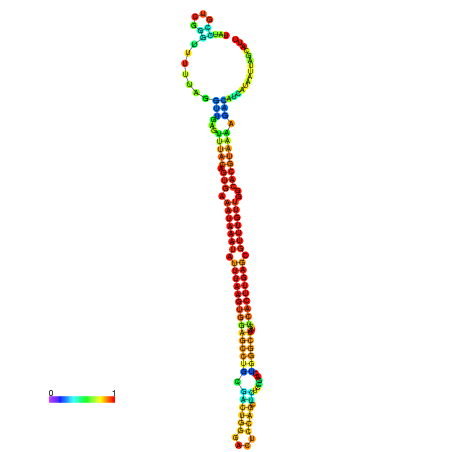
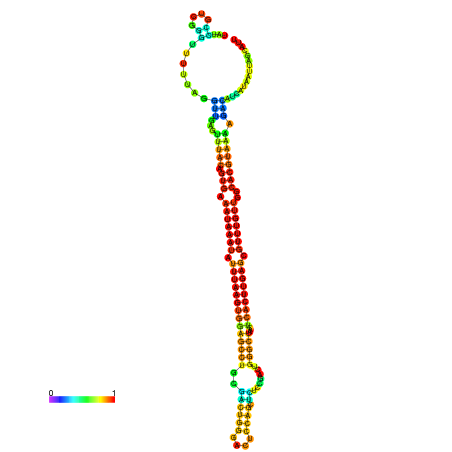
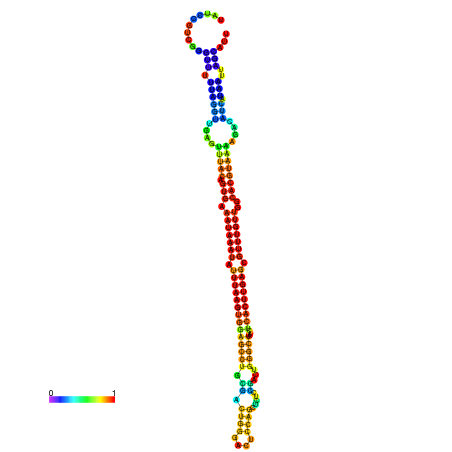
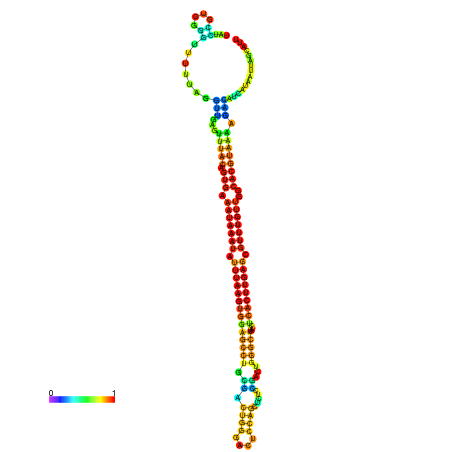
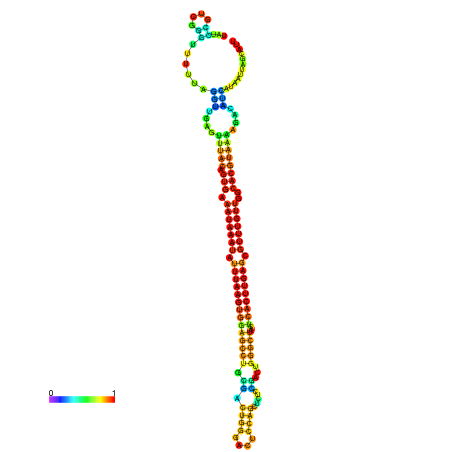
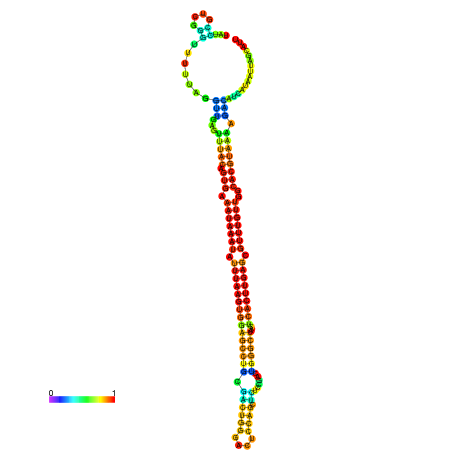
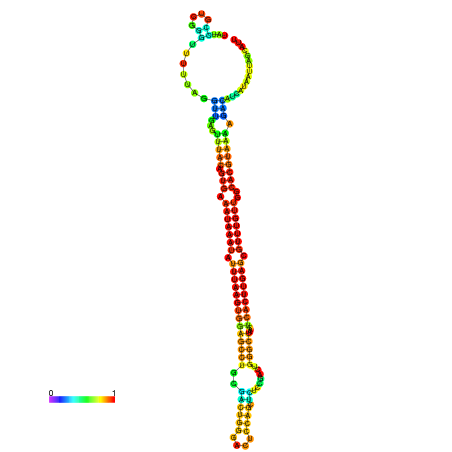
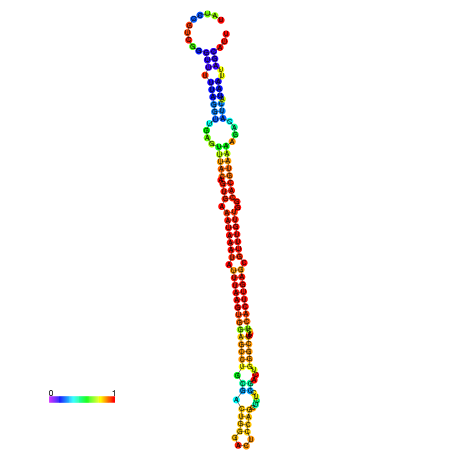
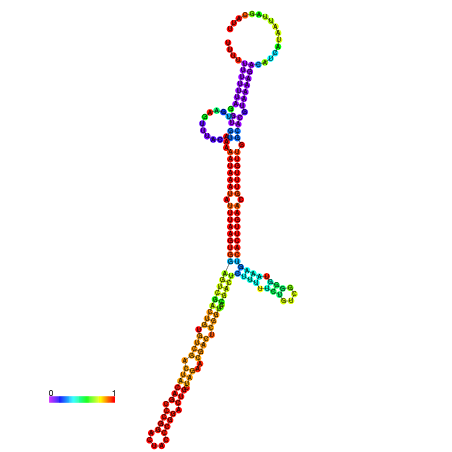
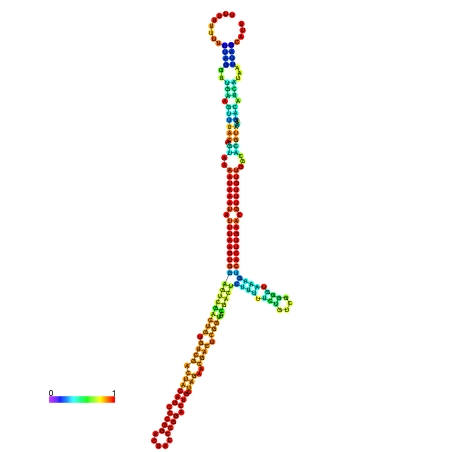
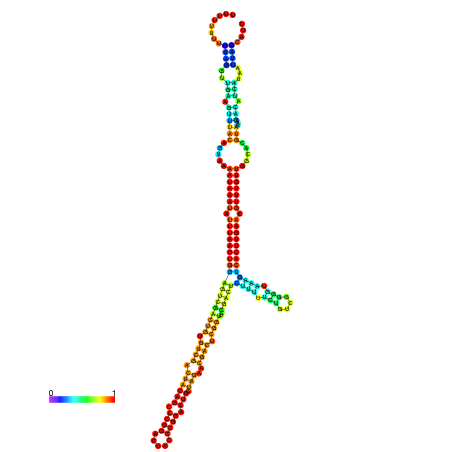
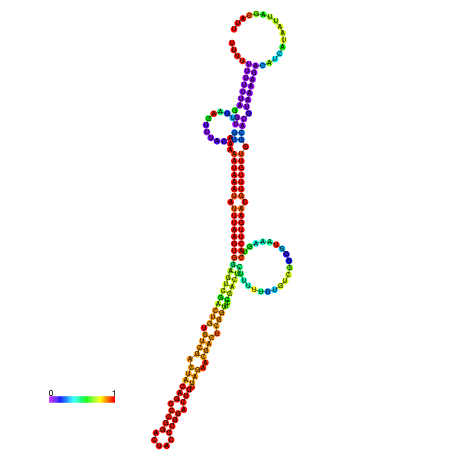
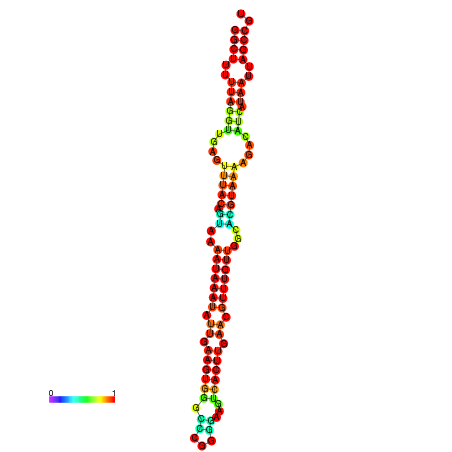
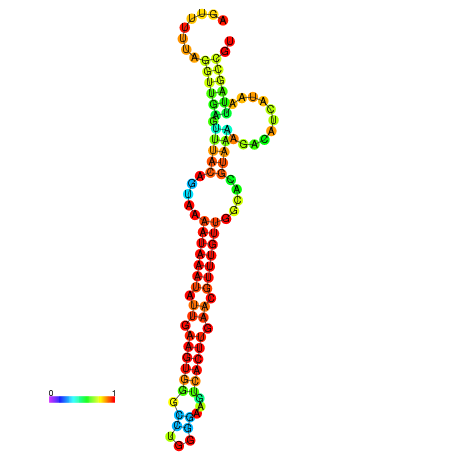
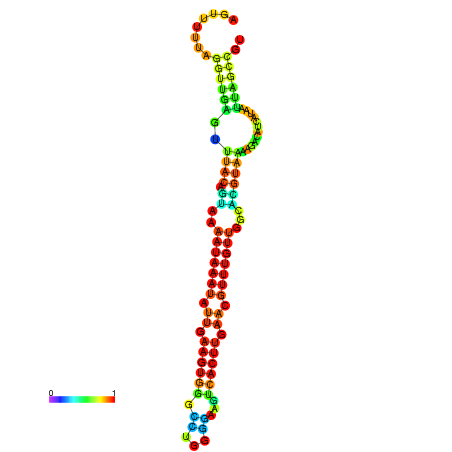
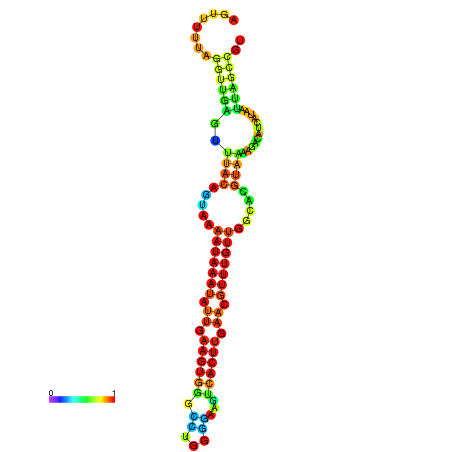
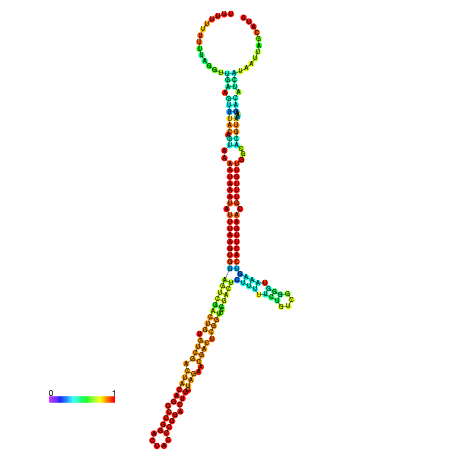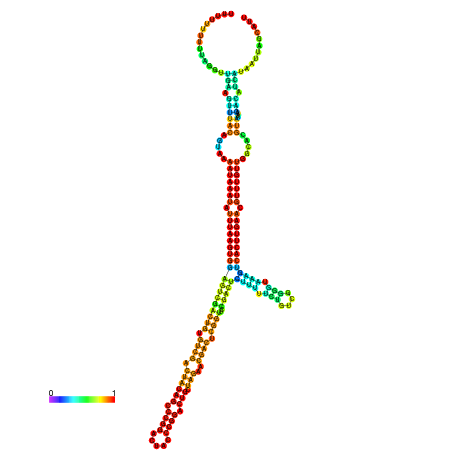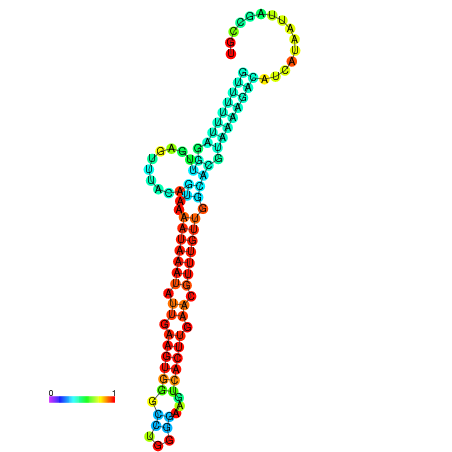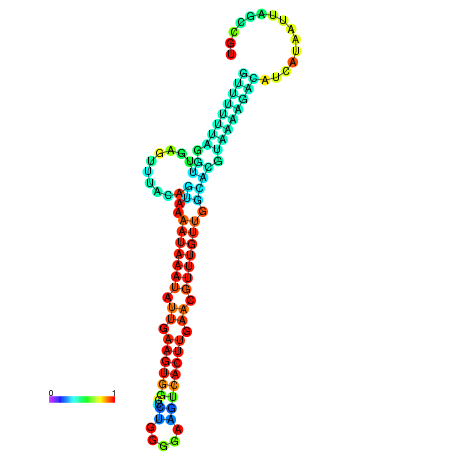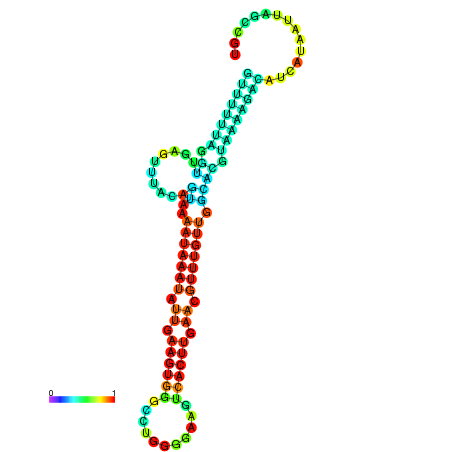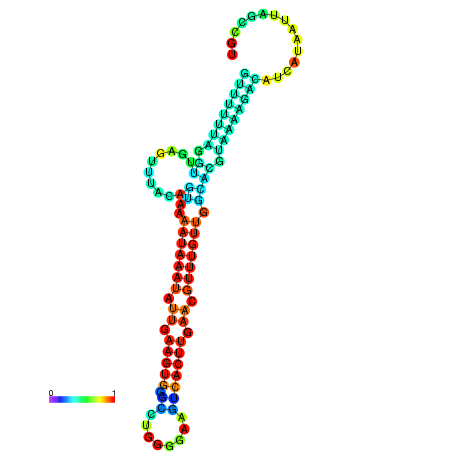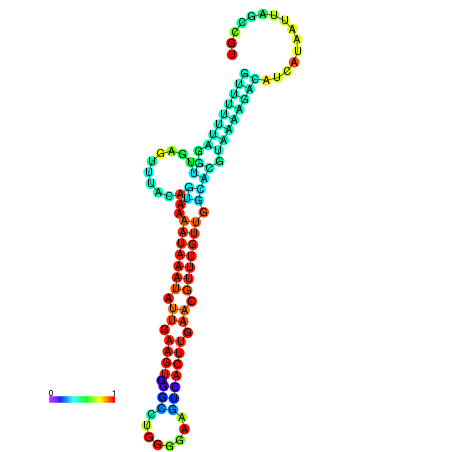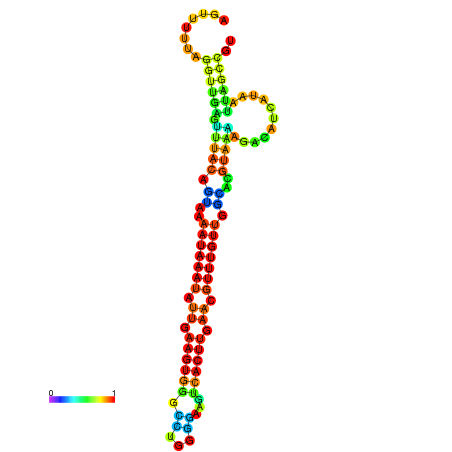| dm3 |
chr3L:13613907-13614035 + |
TAC------------------------GGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGA---------CTT----------------------------------------CAGTCCCTCTGACTGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATT |
| droSim1 |
chr3L:12997848-12997976 + |
TAC------------------------GGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGA---------CTC----------------------------------------CAGTCCCTCTGACTGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATT |
| droSec1 |
super_0:5775586-5775714 + |
TAC------------------------GGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGA---------CTC----------------------------------------CAGTCCCTCTGACTGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATT |
| droYak2 |
chr3L:13706954-13707082 + |
TAC------------------------GGTTCTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGA---------CTC----------------------------------------CAGTCCCTCTGACTGACTGGGGTAAGTCTCTCGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATT |
| droEre2 |
scaffold_4784:13614855-13614983 + |
AAC------------------------GGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGA---------CTC----------------------------------------CAGTCCCTCTGACGGACTGGGGTAACTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATT |
| droAna3 |
scaffold_13337:21827571-21827739 - |
TTC-CGGCACTCCGGCATTCCAGCTC-GGGTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTCTG----CTACTGCC---ACTGCCACTGCCACTGCCACTGCCGCTC------------------------GGGGAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATT |
| dp4 |
chrXR_group8:6818889-6819024 - |
TATCCG------------------TCGGGTTTTTAGGTTGA-GTTTACAGTGAAATAAATATTTAAGTGGAGCCTGCGACTGG---GACTC----------------------------------------CAG-----CTCTCCGACTGGGCTAACTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATT |
| droPer1 |
super_36:488140-488275 + |
TATCCG------------------TCGGGTTTTTAGGTTGA-GTTTACAGTGAAATAAATATTTAAGTGGAGCCTGCGACTGG---GACTC----------------------------------------CAG-----CTCTCCGACTGGGCTAACTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATT |
| droWil1 |
scaffold_180698:607141-607313 - |
TTT------------------------TTTTTTTAGGTTGAAGTTTACAGTAAAATAAATATTTAAGTGGAGTC---GACTGTGTCGACTACAGCCCGGACTACC---CGGACTGCTAGAACGACTCGGTTCCGACTCTTTTTCTGTCGGGGTAAAGTCACTTGAACGTTTGTTGGCACGTAAAAGACATCATAATTAGCATT |
| droVir3 |
scaffold_13049:19817984-19818083 - |
---------------------------GTTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTGAAGTGG---------------------------------------------------------------------------GCCT-GGGGAAGTCACTTGAACGTTTGTTGGCACGTAAAAGACATCATAATTAGCCGT |
| droMoj3 |
scaffold_6680:7513468-7513568 - |
G--------------------------GCTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTGAAGTGGGCCC----------------------------------------------------------------------------GGGGAAGTCACTTGAACGTTTGTTGGCACGTAAAAGACATCATAATTAGCCGT |
| droGri2 |
scaffold_15110:17555344-17555443 + |
---------------------------AGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTGAAGTGG---------------------------------------------------------------------------GCCT-GGGGAAGTCACTTGAACGTTTGTTGGCACGTAAAAGACATCATAATTAGCCGT |