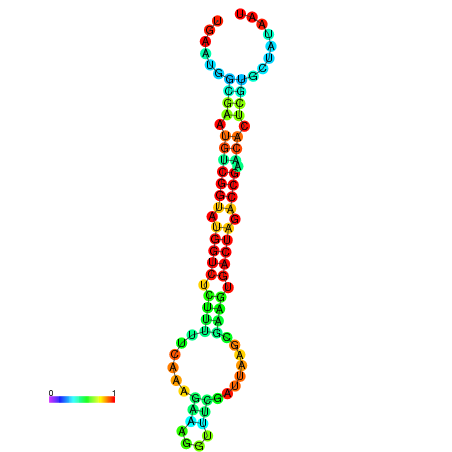
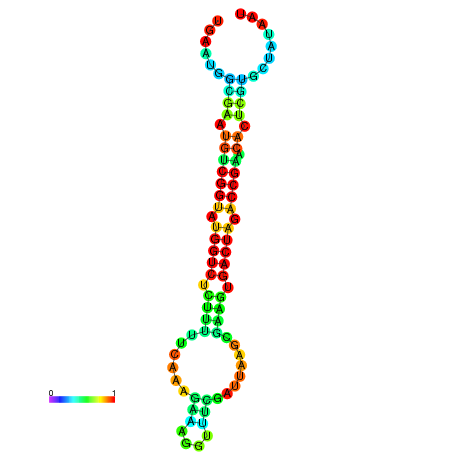
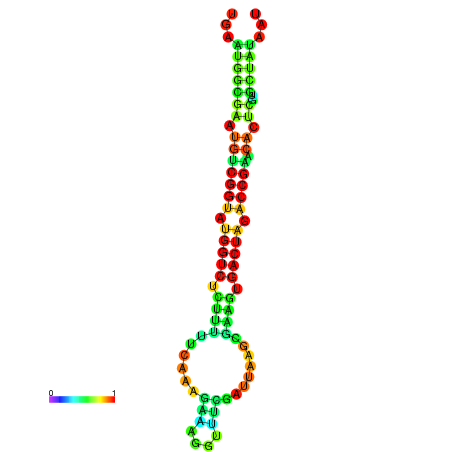

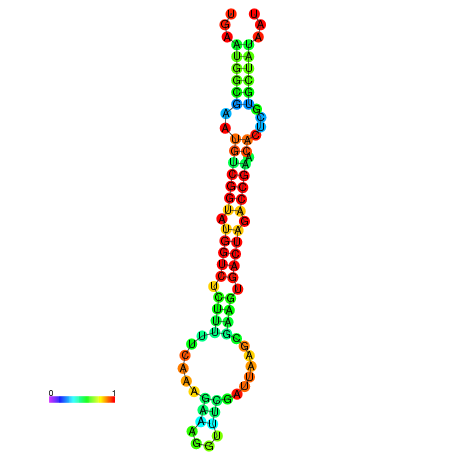
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15548912-15549041 - | TTG-G--------GCAC--------T------CC----------AGTTTTAAAATTGAATGGCGAATGTCGGTATGGTCTCTTTTTCAAA-G-A------AA-------GGTT----TCG--ATTAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATATTCAACATGC-----TCA--G |
| droSim1 | chr2R:14254346-14254476 - | TTG-G--------GCAC--------T------CC----------AGTTTTAAAATTGAATGGCGAATGTCGGTATGGTCTCTTTTTCCAA-AGA------AA-------GGTT----TCG--ATCAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATATTCAACATGC-----TCA--G |
| droSec1 | super_1:13063641-13063770 - | TTG-G--------GCAC--------T------CC----------AGTTTTAAAATTGAATGGCGAATGTCGGTATGGTCTCTTTTTCCAA-T-A------AA-------GGTT----TCG--ATCAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATATTCAACATGC-----TCA--G |
| droYak2 | chr2R:13623749-13623876 + | TAG----------GCAC--------T------CC----------AGTTTTAAAATTGAATGGCGAATGTCGGTATGGTCTCTTTTTCAAA-A--------GA-------GGTT----TCG--ATCAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATATTCAACATGC-----TCA--G |
| droEre2 | scaffold_4845:9749162-9749292 - | TAG----------GCAC--------T------GC----------AGTTTTAAAATTGAATGGCGAATGTCGGTATGGTCTCTTTTTCAAA-G--------AA-------GGTT----TCG--ATCGA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATATTCAACATGC-----ACACTG |
| droAna3 | scaffold_13266:9731344-9731477 + | TTT-A--------GCAT--------C------AG----------AATTTTAAAATTGAATGGCGATTGTCGGTATGGTCGCTTTTTCACA-A-A------AAAATT---GGTT----ACC--AACAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATGTTCATCAAGC-----TAA--A |
| dp4 | chr3:12677574-12677700 - | AAA-A--------GAAG--------A------CA----------AATTTTAAAATTAAATGGCGATTGTCGGTTTGGTCGCTTTTTACCA-----------G-------GGTT----CCG--ATCAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAGAATGTTCAACATGG-----CCA--A |
| droPer1 | super_2:8905966-8906092 + | AAA-A--------GAAG--------A------CA----------AATTTTAAAATTAAATGGCGATTGTCGGTTTGGTCGCTTTTTACCA-----------G-------GGTT----CCG--ATCAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAGAATGTTCAACATGG-----CCA--A |
| droWil1 | scaffold_180701:1651730-1651882 - | TAT----------GCAA--------T------TG-AAATT----ACTTTTAAAATTGAATGGCGATTGTCGGTATAGTCTCTTTTTCCAA-A--------AATCTTGTTTGTTTTTGCCA--ATCTAATAAGAAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATGTTCAACAAGCAACAACCA--A |
| droVir3 | scaffold_12875:4112084-4112243 + | GTG-----TTCCAGTATTCCAGTGTTTCAATTCCAATATTCCAGTATTTTAAAATTAAATGGCGATTGTCGGTATGGTCGCTTTTTCAAA-C--------AA-------TATA----CCATTATC--TATATAAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATGTTCAACGTAT-----ATA--A |
| droMoj3 | scaffold_6496:20936771-20936914 - | TCC-A--------GTGT--------T------CGAAAATTATAAAATTTTAAAATTAAATGGCGATTGTCGGTATGGTCTCTTTTTCAACAT-ATACACCAA-------ACAT----T------C--TATATAAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATGTTCAACAATT-----TCA--A |
| droGri2 | scaffold_15112:2108802-2108953 - | TGTGTGTGTT---GCAG--------T-CAAACCCAG----CCAGTATTTTAAAATTAAATGGCGATTGTCGGTATGGTCGCTTTTGAACC-T-AAACTC------T---ATAT----ACA--ATA--TATATAAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATGTTCAACATAG-----GCA--T |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15548912-15549041 - | TTG-G--------GCAC--------T------CC----------AGTTTTAAAATTGAATGGCGAATGTCGGTATGGTCTCTTTTTCAAA-G-A------AA-------GGTT----TCG--ATTAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATATTCAACATGC-----TCA--G |
| droSim1 | chr2R:14254346-14254476 - | TTG-G--------GCAC--------T------CC----------AGTTTTAAAATTGAATGGCGAATGTCGGTATGGTCTCTTTTTCCAA-AGA------AA-------GGTT----TCG--ATCAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATATTCAACATGC-----TCA--G |
| droSec1 | super_1:13063641-13063770 - | TTG-G--------GCAC--------T------CC----------AGTTTTAAAATTGAATGGCGAATGTCGGTATGGTCTCTTTTTCCAA-T-A------AA-------GGTT----TCG--ATCAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATATTCAACATGC-----TCA--G |
| droYak2 | chr2R:13623749-13623876 + | TAG----------GCAC--------T------CC----------AGTTTTAAAATTGAATGGCGAATGTCGGTATGGTCTCTTTTTCAAA-A--------GA-------GGTT----TCG--ATCAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATATTCAACATGC-----TCA--G |
| droEre2 | scaffold_4845:9749162-9749292 - | TAG----------GCAC--------T------GC----------AGTTTTAAAATTGAATGGCGAATGTCGGTATGGTCTCTTTTTCAAA-G--------AA-------GGTT----TCG--ATCGA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATATTCAACATGC-----ACACTG |
| droAna3 | scaffold_13266:9731344-9731477 + | TTT-A--------GCAT--------C------AG----------AATTTTAAAATTGAATGGCGATTGTCGGTATGGTCGCTTTTTCACA-A-A------AAAATT---GGTT----ACC--AACAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATGTTCATCAAGC-----TAA--A |
| dp4 | chr3:12677574-12677700 - | AAA-A--------GAAG--------A------CA----------AATTTTAAAATTAAATGGCGATTGTCGGTTTGGTCGCTTTTTACCA-----------G-------GGTT----CCG--ATCAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAGAATGTTCAACATGG-----CCA--A |
| droPer1 | super_2:8905966-8906092 + | AAA-A--------GAAG--------A------CA----------AATTTTAAAATTAAATGGCGATTGTCGGTTTGGTCGCTTTTTACCA-----------G-------GGTT----CCG--ATCAA---GCGAAGTGACTAGACCGAACACTCGTGCTATAATTTTAGAATGTTCAACATGG-----CCA--A |
| droWil1 | scaffold_180701:1651730-1651882 - | TAT----------GCAA--------T------TG-AAATT----ACTTTTAAAATTGAATGGCGATTGTCGGTATAGTCTCTTTTTCCAA-A--------AATCTTGTTTGTTTTTGCCA--ATCTAATAAGAAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATGTTCAACAAGCAACAACCA--A |
| droVir3 | scaffold_12875:4112084-4112243 + | GTG-----TTCCAGTATTCCAGTGTTTCAATTCCAATATTCCAGTATTTTAAAATTAAATGGCGATTGTCGGTATGGTCGCTTTTTCAAA-C--------AA-------TATA----CCATTATC--TATATAAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATGTTCAACGTAT-----ATA--A |
| droMoj3 | scaffold_6496:20936771-20936914 - | TCC-A--------GTGT--------T------CGAAAATTATAAAATTTTAAAATTAAATGGCGATTGTCGGTATGGTCTCTTTTTCAACAT-ATACACCAA-------ACAT----T------C--TATATAAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATGTTCAACAATT-----TCA--A |
| droGri2 | scaffold_15112:2108802-2108953 - | TGTGTGTGTT---GCAG--------T-CAAACCCAG----CCAGTATTTTAAAATTAAATGGCGATTGTCGGTATGGTCGCTTTTGAACC-T-AAACTC------T---ATAT----ACA--ATA--TATATAAAGTGACTAGACCGAACACTCGTGCTATAATTTTAAAATGTTCAACATAG-----GCA--T |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-21.1, p-value=0.009901 | dG=-21.1, p-value=0.009901 | dG=-21.0, p-value=0.009901 | dG=-21.0, p-value=0.009901 | dG=-20.9, p-value=0.009901 |
|---|---|---|---|---|
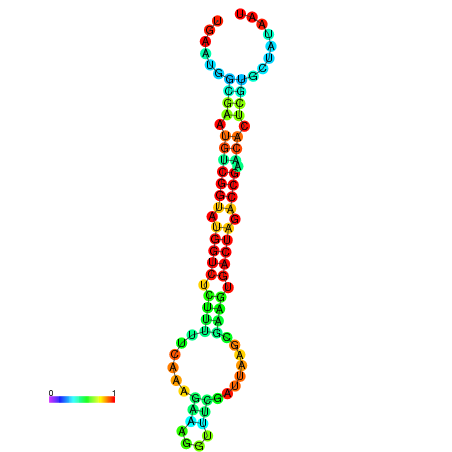 |
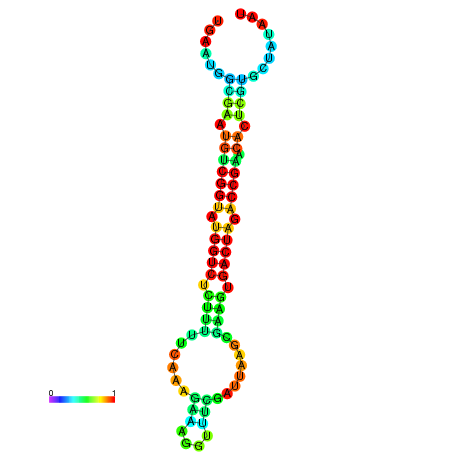 |
 |
 |
 |
| dG=-21.7, p-value=0.009901 | dG=-21.7, p-value=0.009901 | dG=-21.6, p-value=0.009901 | dG=-21.6, p-value=0.009901 | dG=-21.5, p-value=0.009901 |
|---|---|---|---|---|
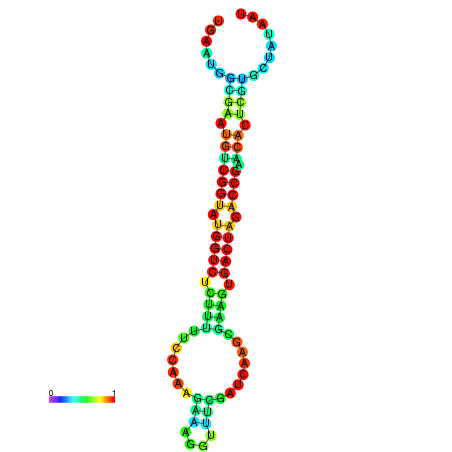 |
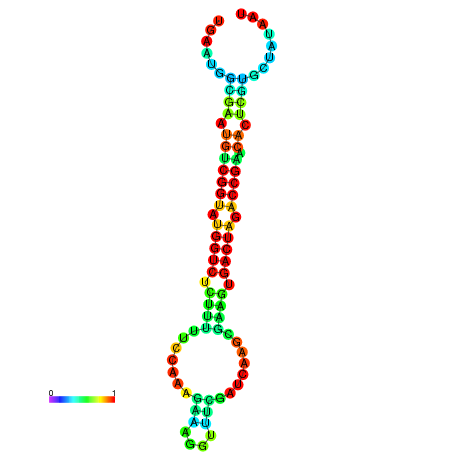 |
 |
 |
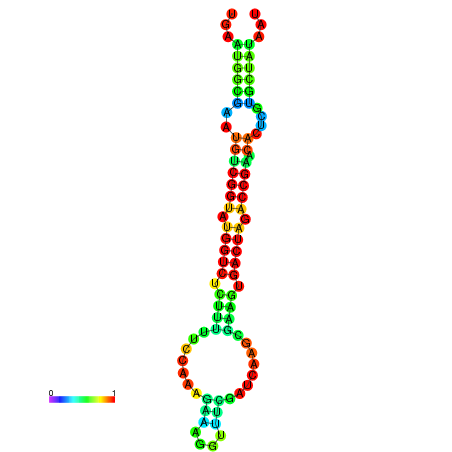 |
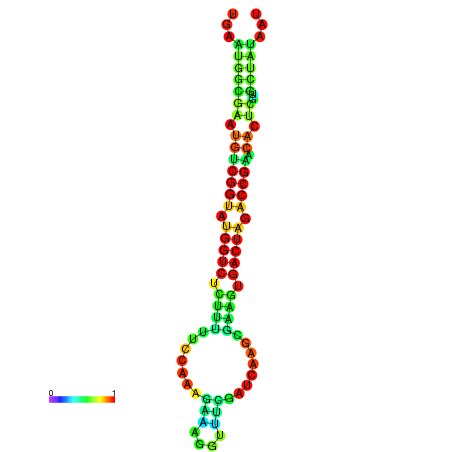
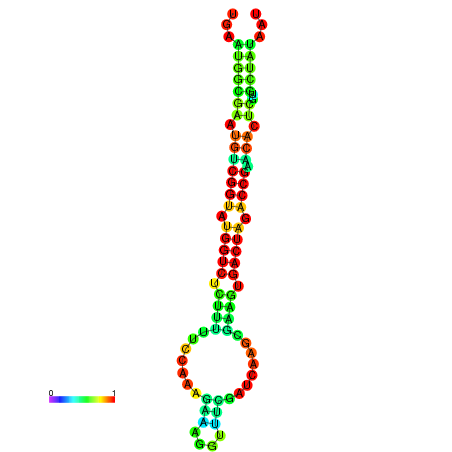
| dG=-20.1, p-value=0.009901 | dG=-20.1, p-value=0.009901 | dG=-20.1, p-value=0.009901 | dG=-20.1, p-value=0.009901 | dG=-20.0, p-value=0.009901 |
|---|---|---|---|---|
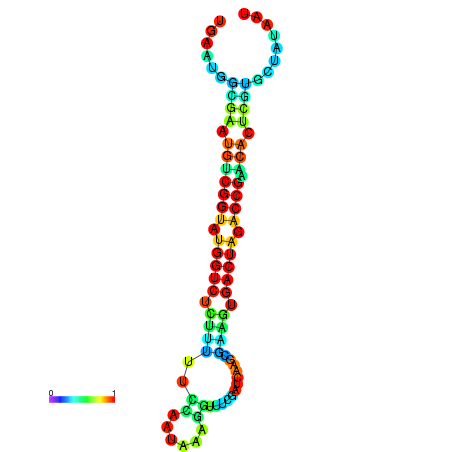 |
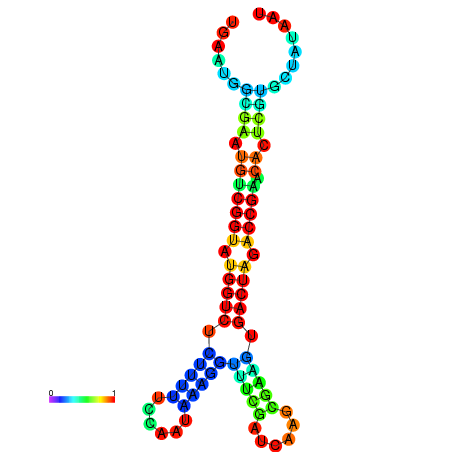 |
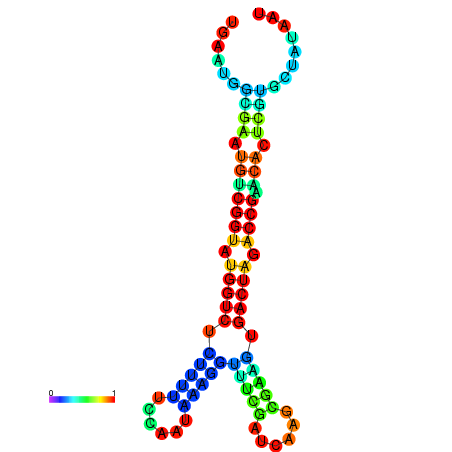 |
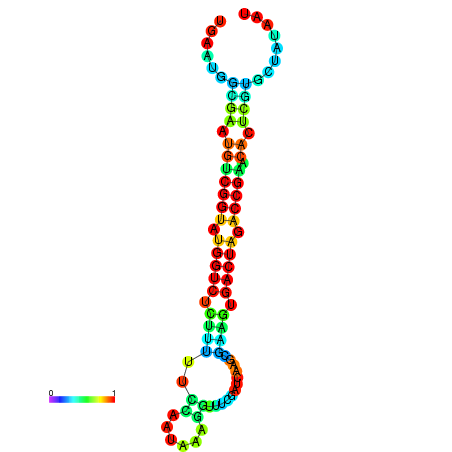 |
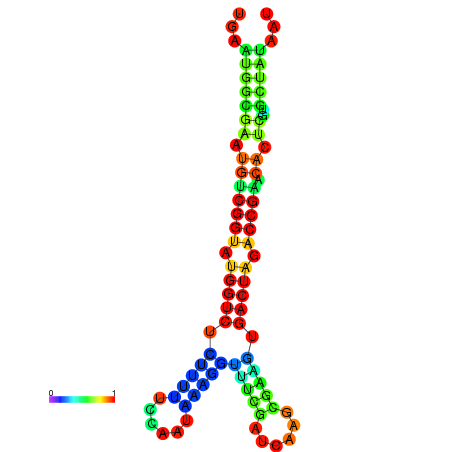 |
| dG=-20.8, p-value=0.009901 | dG=-20.8, p-value=0.009901 | dG=-20.7, p-value=0.009901 | dG=-20.7, p-value=0.009901 | dG=-20.6, p-value=0.009901 |
|---|---|---|---|---|
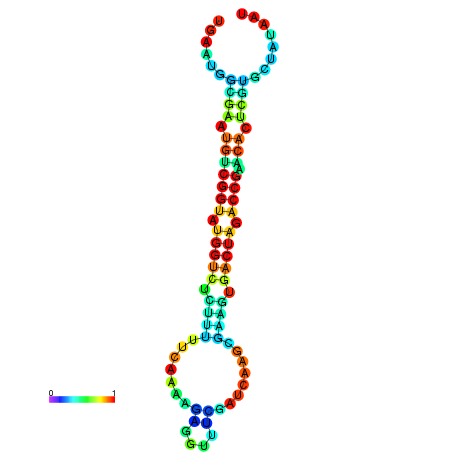 |
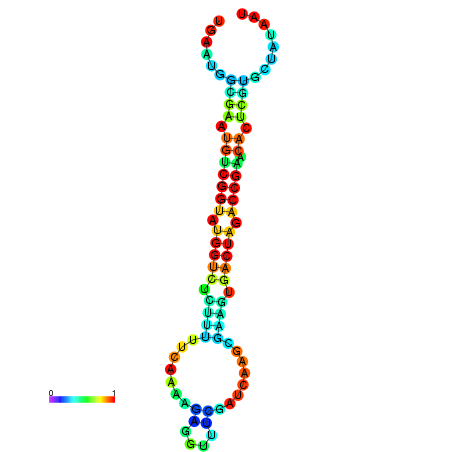 |
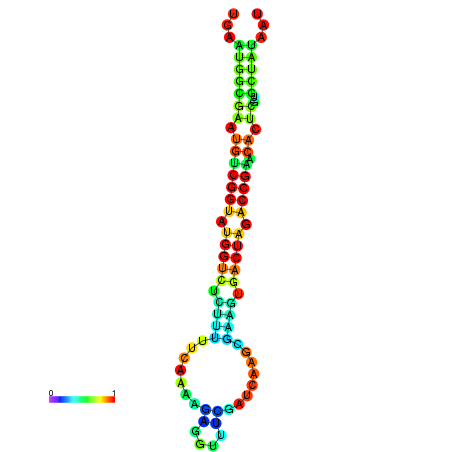 |
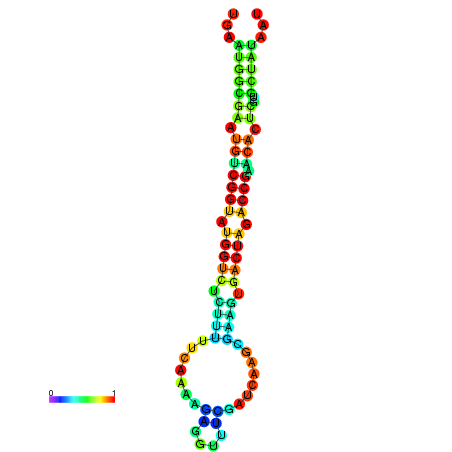 |
 |
| dG=-22.1, p-value=0.009901 | dG=-22.1, p-value=0.009901 | dG=-22.0, p-value=0.009901 | dG=-22.0, p-value=0.009901 | dG=-21.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-24.2, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
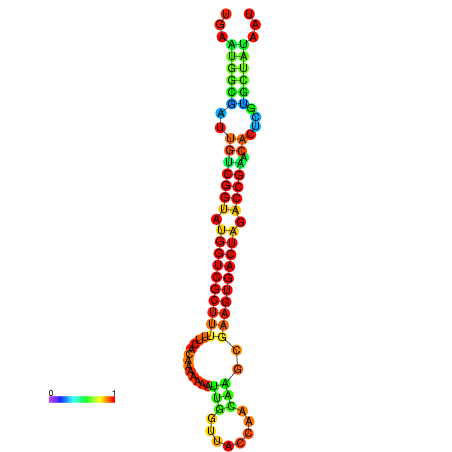 |
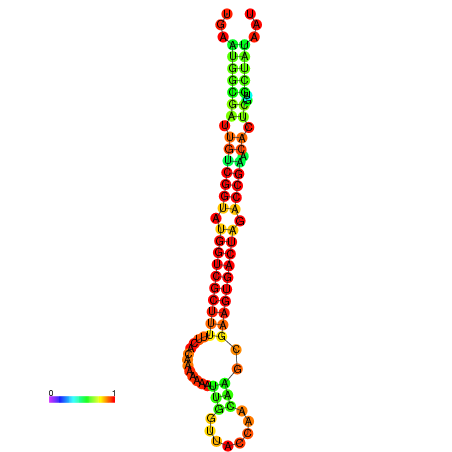
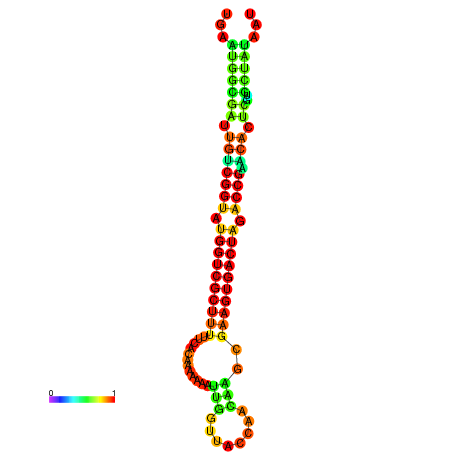
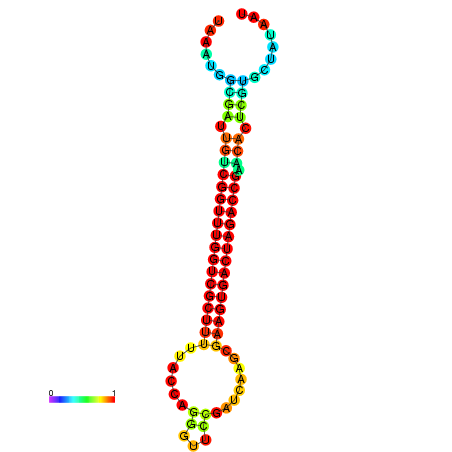
| dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
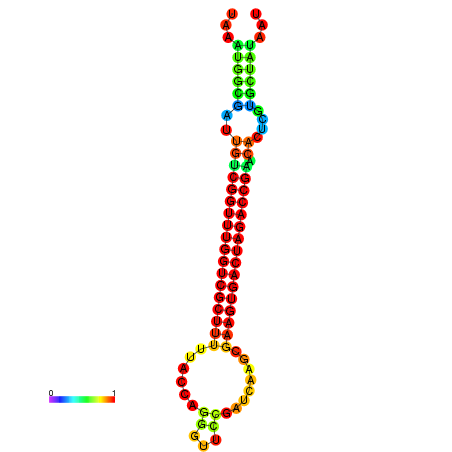 |
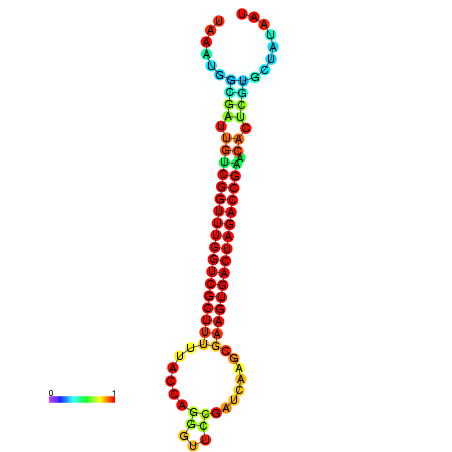
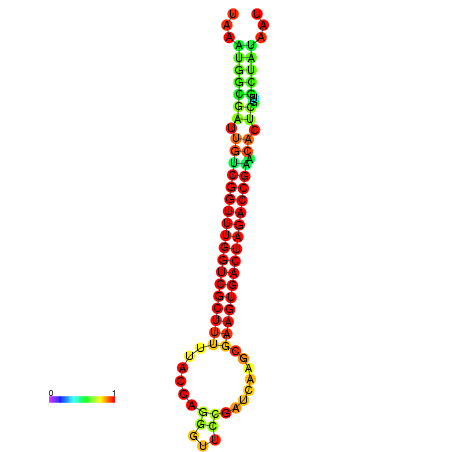
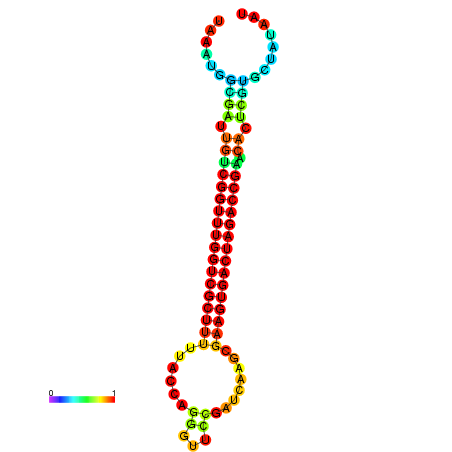
| dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
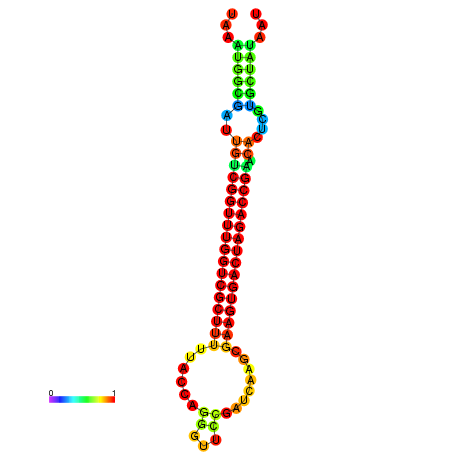 |
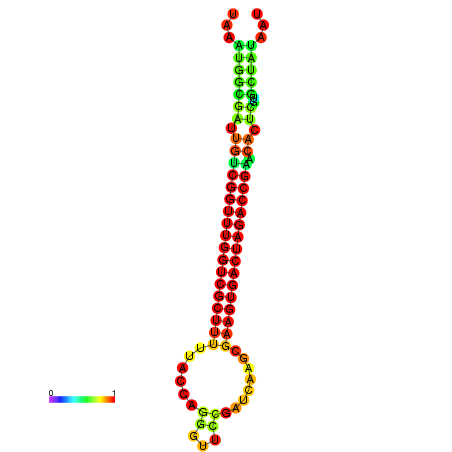
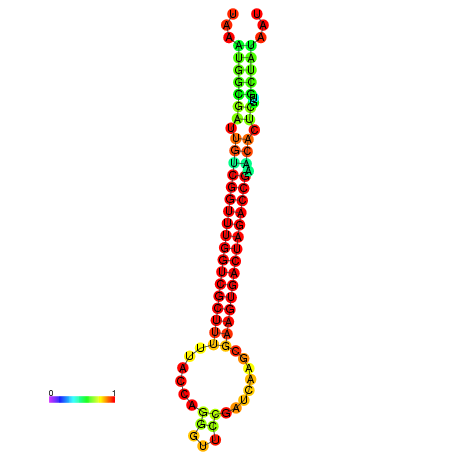
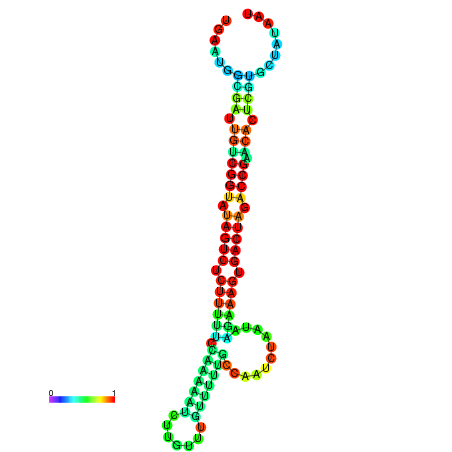
| dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
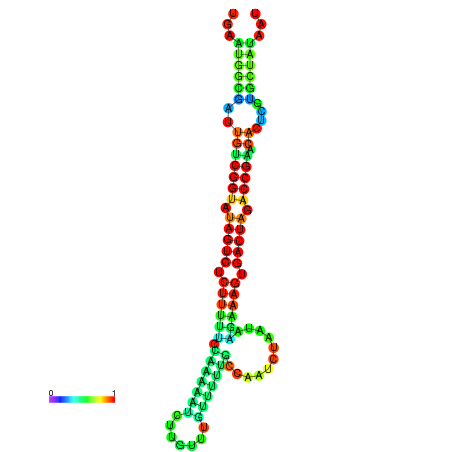 |
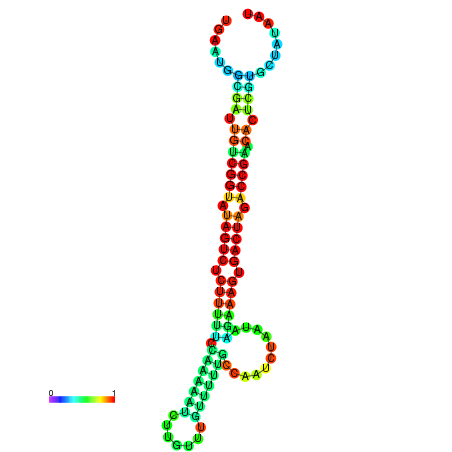
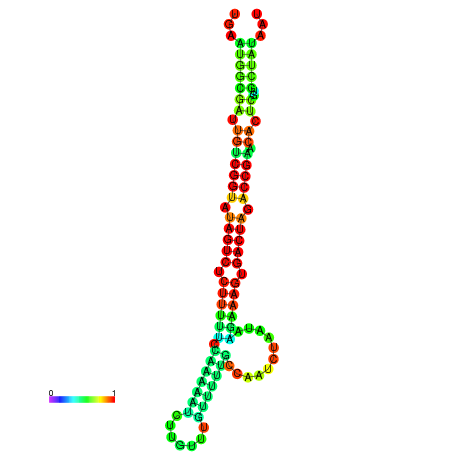
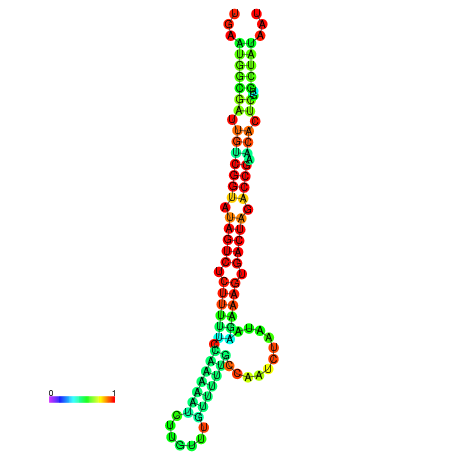
| dG=-23.9, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.7, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
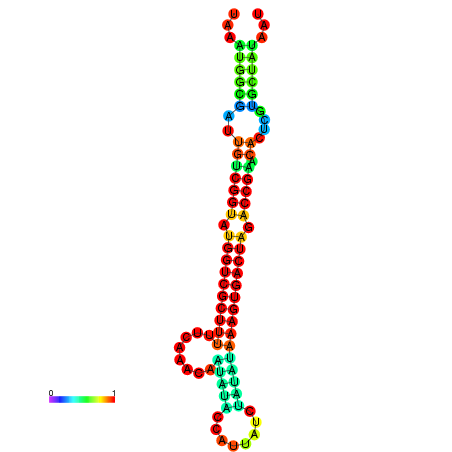 |
| dG=-19.6, p-value=0.009901 | dG=-19.6, p-value=0.009901 | dG=-19.5, p-value=0.009901 | dG=-19.5, p-value=0.009901 | dG=-19.4, p-value=0.009901 |
|---|---|---|---|---|
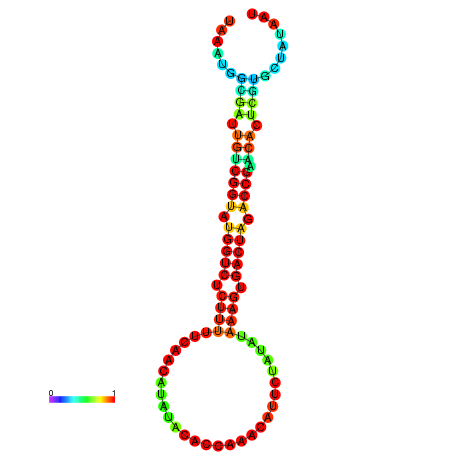 |
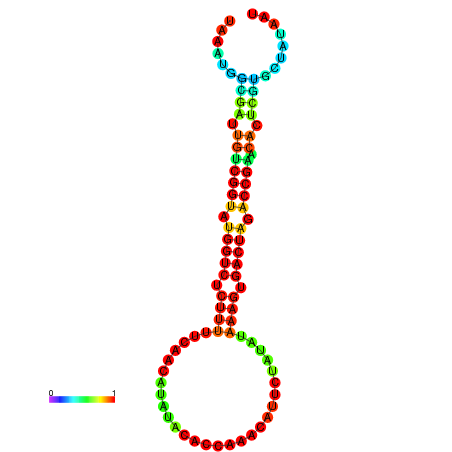 |
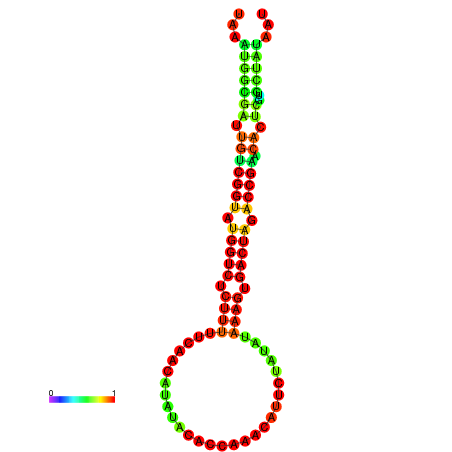 |
 |
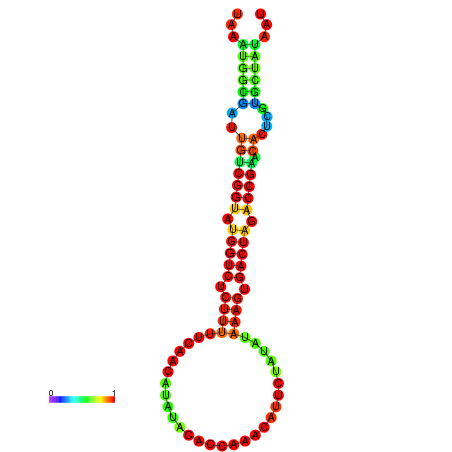 |
| dG=-23.4, p-value=0.009901 | dG=-23.4, p-value=0.009901 | dG=-23.4, p-value=0.009901 | dG=-23.4, p-value=0.009901 | dG=-23.3, p-value=0.009901 |
|---|---|---|---|---|
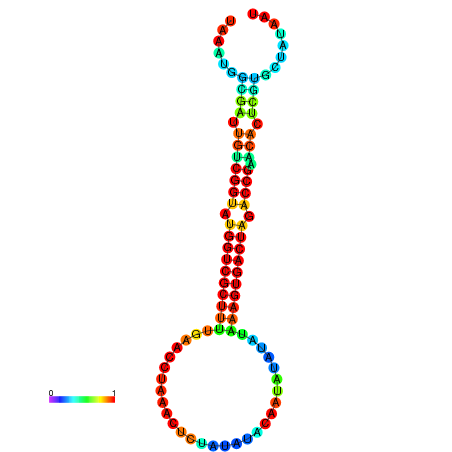 |
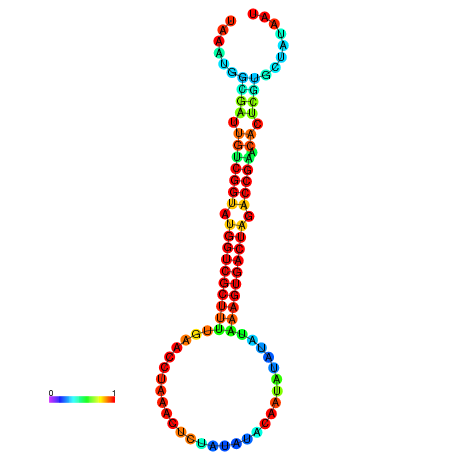 |
 |
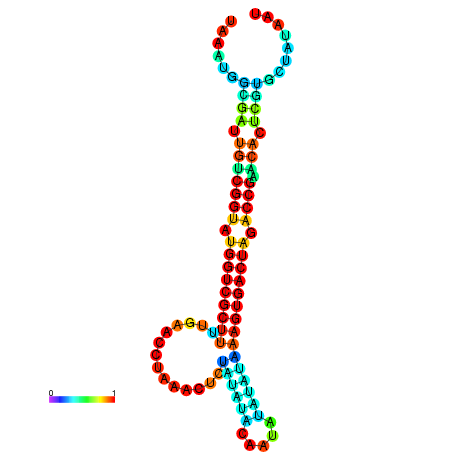 |
 |
Generated: 03/07/2013 at 05:06 PM