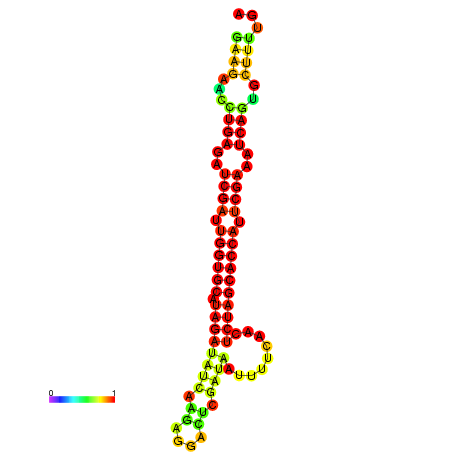
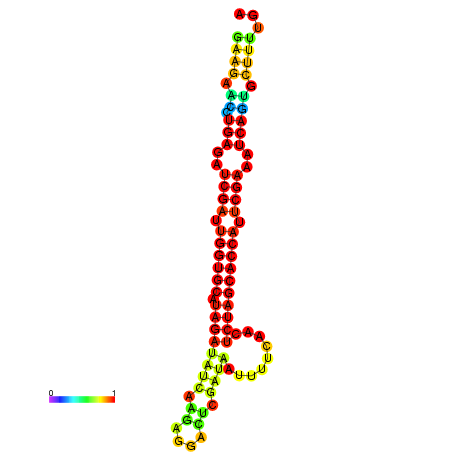
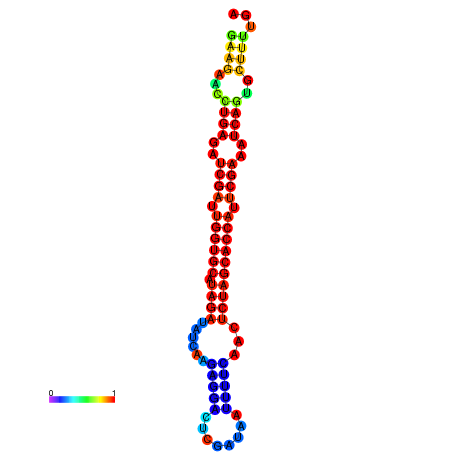
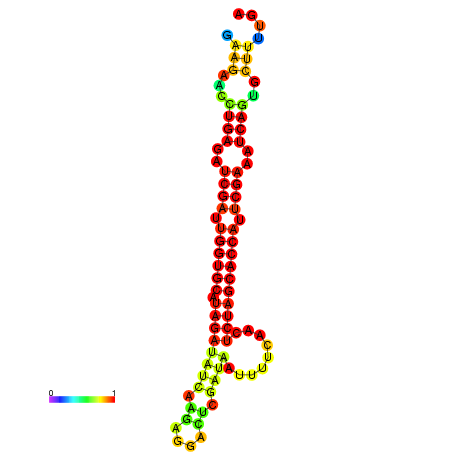
| TCTTACAATTACAGTTCGAATCGAAGAACTGAGATCGATTGGTGCATAGATATCAAGAGGACTCGCTAATTTTCAACTCTAGCACCATTCGAAATCAGTGCTTTTGATGAGAACCATTCAACAGA----- | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ...............................................................................TAGCACCATTCGAAATCAGTGCT............................ | 23 | 1 | 24878.00 | 24878 | 3 | 24875 |
| ...............................................................................TAGCACCATTCGAAATCAGTGC............................. | 22 | 1 | 12336.00 | 12336 | 1 | 12335 |
| ...............................................................................TAGCACCATTCGAAATCAGTG.............................. | 21 | 1 | 1854.00 | 1854 | 0 | 1854 |
| ...............................................................................TAGCACCATTCGAAATCAGT............................... | 20 | 1 | 1064.00 | 1064 | 0 | 1064 |
| ...........................ACTGAGATCGATTGGTGCATAGA................................................................................ | 23 | 1 | 426.00 | 426 | 2 | 424 |
| ...............................................................................TAGCACCATTCGAAATCAG................................ | 19 | 1 | 94.00 | 94 | 0 | 94 |
| ...........................ACTGAGATCGATTGGTGCATAG................................................................................. | 22 | 1 | 73.00 | 73 | 0 | 73 |
| ..........................AACTGAGATCGATTGGTGCATAGA................................................................................ | 24 | 1 | 55.00 | 55 | 0 | 55 |
| ...............................................................................TAGCACCATTCGAAATCAGTGCTT........................... | 24 | 1 | 44.00 | 44 | 0 | 44 |
| ..........................AACTGAGATCGATTGGTGCATAG................................................................................. | 23 | 1 | 44.00 | 44 | 0 | 44 |
| ..............................................................................CTAGCACCATTCGAAATCAGTGCT............................ | 24 | 1 | 34.00 | 34 | 0 | 34 |
| ................................................................................AGCACCATTCGAAATCAGTGCT............................ | 22 | 1 | 33.00 | 33 | 0 | 33 |
| ..........................AACTGAGATCGATTGGTGCAT................................................................................... | 21 | 1 | 33.00 | 33 | 0 | 33 |
| ..............................................................................CTAGCACCATTCGAAATCAGTGC............................. | 23 | 1 | 28.00 | 28 | 0 | 28 |
| ...........................ACTGAGATCGATTGGTGCAT................................................................................... | 20 | 1 | 22.00 | 22 | 0 | 22 |
| ...........................ACTGAGATCGATTGGTGCATA.................................................................................. | 21 | 1 | 17.00 | 17 | 0 | 17 |
| ................................................................................AGCACCATTCGAAATCAGTGC............................. | 21 | 1 | 17.00 | 17 | 0 | 17 |
| .................................................................................GCACCATTCGAAATCAGTGCT............................ | 21 | 1 | 13.00 | 13 | 0 | 13 |
| ..........................AACTGAGATCGATTGGTGCATA.................................................................................. | 22 | 1 | 9.00 | 9 | 0 | 9 |
| ..........................AACTGAGATCGATTGGTGCA.................................................................................... | 20 | 1 | 6.00 | 6 | 0 | 6 |
| ............................CTGAGATCGATTGGTGCATAGA................................................................................ | 22 | 1 | 6.00 | 6 | 0 | 6 |
| .................................................................................GCACCATTCGAAATCAGTGC............................. | 20 | 1 | 6.00 | 6 | 0 | 6 |
| ..............................................................................CTAGCACCATTCGAAATCAGTG.............................. | 22 | 1 | 5.00 | 5 | 0 | 5 |
| ..................................................TATCAAGAGGACTCGCTAATT........................................................... | 21 | 1 | 4.00 | 4 | 0 | 4 |
| .............................TGAGATCGATTGGTGCATAGA................................................................................ | 21 | 1 | 4.00 | 4 | 0 | 4 |
| ..................................................TATCAAGAGGACTCGCTAATTTTC........................................................ | 24 | 1 | 3.00 | 3 | 0 | 3 |