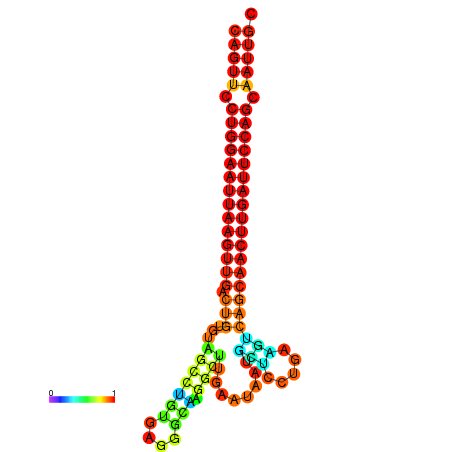
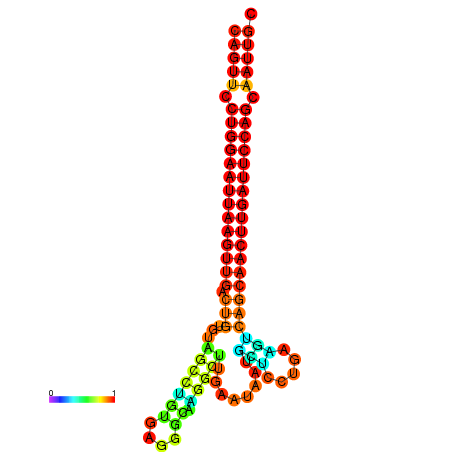
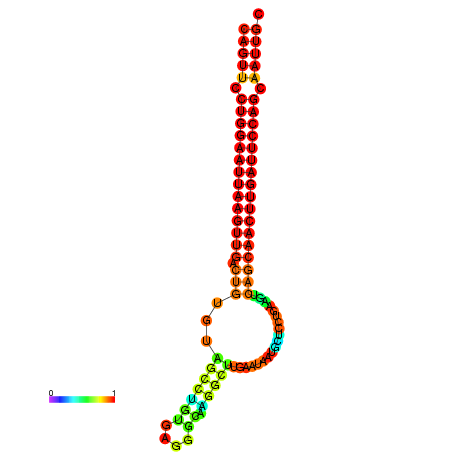
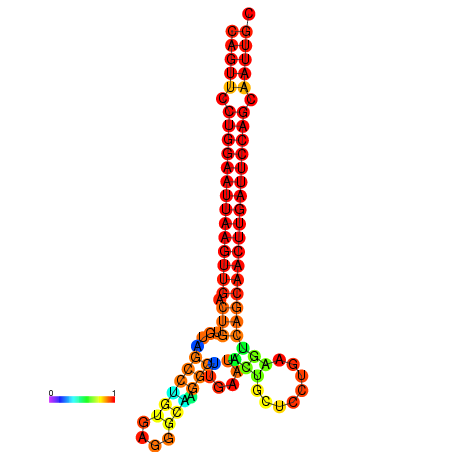
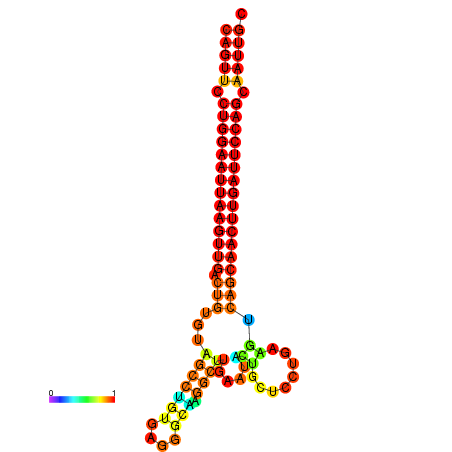
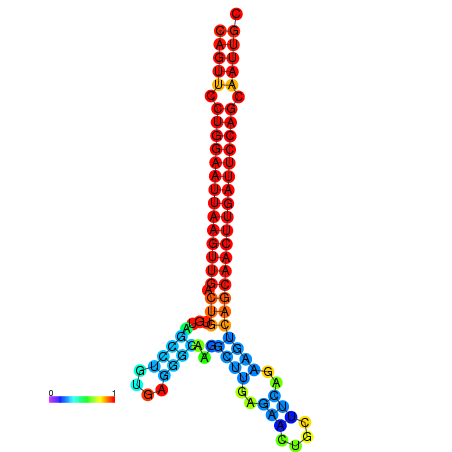
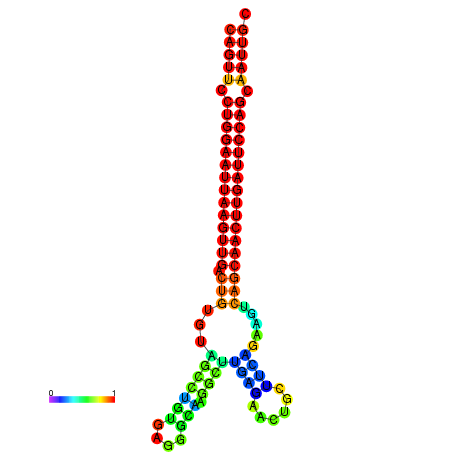
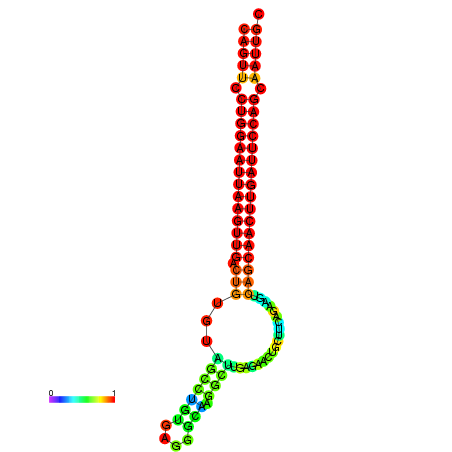
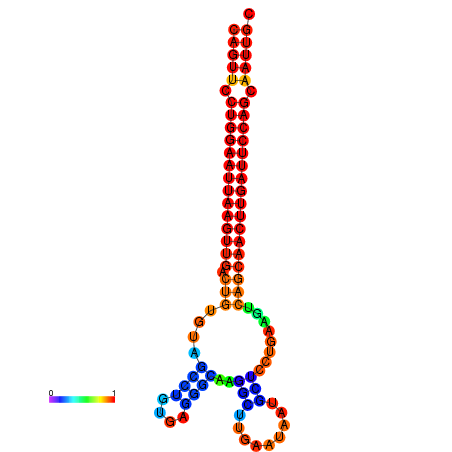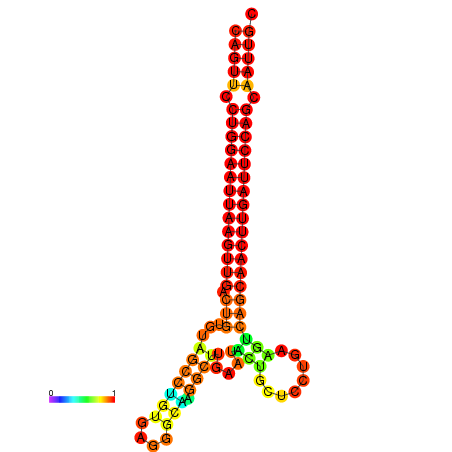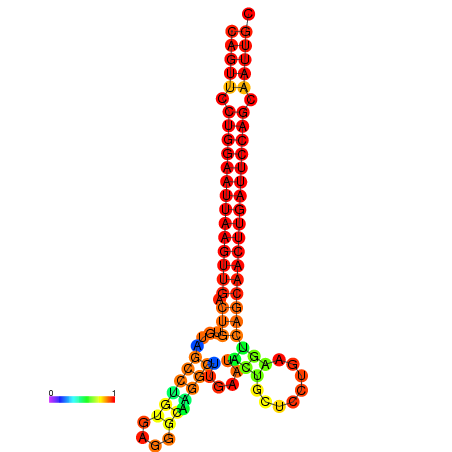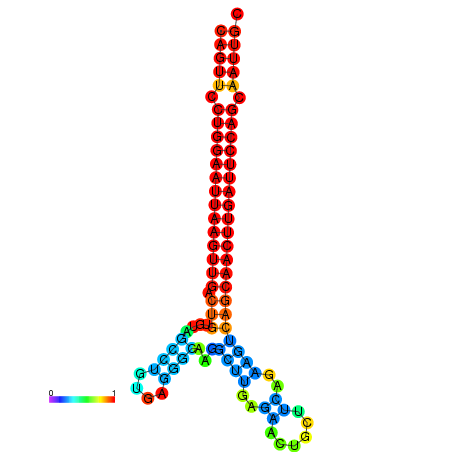| ATCAGAGGCAATTGCGTTGCAGTTCCTGGAATTAAGTTGACTGTGTAGCCTGGGAAGGCAAGGCTTGAGCACTGCTTCTGAAGTCAGCAACTTGATTCCAGCAATTGCGGCCCAAG----CACAACGCTGAATGGTGCTCAAAATAC | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ..................................................................................GTCAGCAACTTGATTCCAGCAA........................................... | 22 | 1 | 1410.00 | 1410 | 244 | 1166 |
| ........................CCTGGAATTAAGTTGACTGTGTAGCC................................................................................................. | 26 | 1 | 994.00 | 994 | 127 | 867 |
| ..................................................................................GTCAGCAACTTGATTCCAGCA............................................ | 21 | 1 | 981.00 | 981 | 53 | 928 |
| ........................CCTGGAATTAAGTTGACTGTGTAGC.................................................................................................. | 25 | 1 | 490.00 | 490 | 67 | 423 |
| ........................CCTGGAATTAAGTTGACTGTGTAG................................................................................................... | 24 | 1 | 204.00 | 204 | 3 | 201 |
| .....AGGCAATTGCGTTGCAGTT........................................................................................................................... | 19 | 1 | 157.00 | 157 | 6 | 151 |
| ........................CCTGGAATTAAGTTGACTGTGT..................................................................................................... | 22 | 1 | 93.00 | 93 | 1 | 92 |
| ........................CCTGGAATTAAGTTGACTGTG...................................................................................................... | 21 | 1 | 59.00 | 59 | 0 | 59 |
| ........................CCTGGAATTAAGTTGACTGTGTA.................................................................................................... | 23 | 1 | 50.00 | 50 | 1 | 49 |
| ..................................................................................GTCAGCAACTTGATTCCAGC............................................. | 20 | 1 | 31.00 | 31 | 0 | 31 |
| ........................CCTGGAATTAAGTTGACTGTGTAGCCT................................................................................................ | 27 | 1 | 21.00 | 21 | 0 | 21 |
| ........................CCTGGAATTAAGTTGACTGT....................................................................................................... | 20 | 1 | 8.00 | 8 | 0 | 8 |
| .....................................................................................AGCAACTTGATTCCAGCAA........................................... | 19 | 1 | 8.00 | 8 | 3 | 5 |
| .....................................................................................AGCAACTTGATTCCAGCA............................................ | 18 | 1 | 7.00 | 7 | 1 | 6 |
| ..................................................................................GTCAGCAACTTGATTCCAG.............................................. | 19 | 1 | 4.00 | 4 | 0 | 4 |
| ........................CCTGGAATTAAGTTGACTG........................................................................................................ | 19 | 1 | 3.00 | 3 | 0 | 3 |
| .........................CTGGAATTAAGTTGACTGTGTAGC.................................................................................................. | 24 | 1 | 3.00 | 3 | 1 | 2 |
| ...................................................................................TCAGCAACTTGATTCCAGCAA........................................... | 21 | 1 | 2.00 | 2 | 1 | 1 |
| ...AGAGGCAATTGCGTTGCAGTT........................................................................................................................... | 21 | 1 | 2.00 | 2 | 1 | 1 |
| .........................CTGGAATTAAGTTGACTGTGTAG................................................................................................... | 23 | 1 | 2.00 | 2 | 0 | 2 |
| ..CAGAGGCAATTGCGTTGCAGTT........................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 1 |
| .........................CTGGAATTAAGTTGACTGTGTAGCC................................................................................................. | 25 | 1 | 1.00 | 1 | 0 | 1 |
| ....................AGTTCCTGGAATTAAGTTGA........................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 |
| ................................................................................AAGTCAGCAACTTGATTCCAGCA............................................ | 23 | 1 | 1.00 | 1 | 1 | 0 |
| .....AGGCAATTGCGTTGCAGT............................................................................................................................ | 18 | 1 | 1.00 | 1 | 0 | 1 |
| ....................................................................................CAGCAACTTGATTCCAGCA............................................ | 19 | 1 | 1.00 | 1 | 0 | 1 |
| .....AGGCAATTGCGTTGCAGTTC.......................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 |