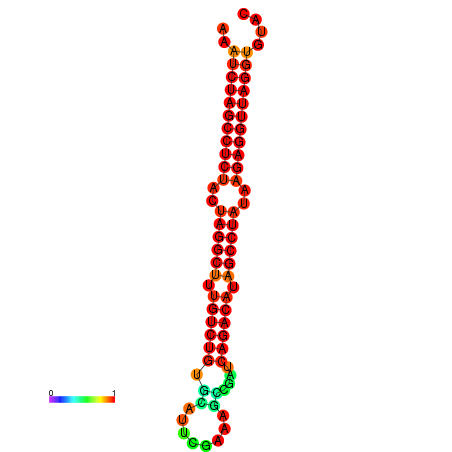
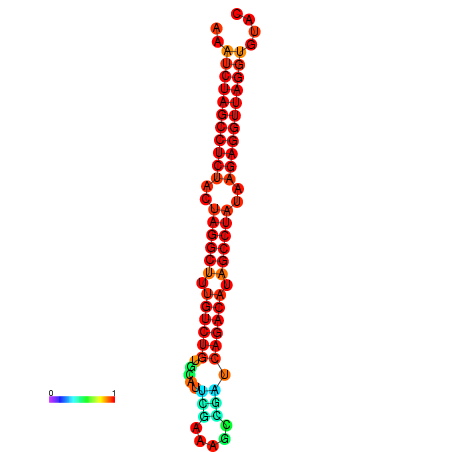
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:3251007-3251133 + | CCGTTCTTT---------AACTATAGTTTCCTTCTAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCGAA-AGCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGCGAACAATCAGC------------------GAAAACGGA |
| droSim1 | chr3L:2784381-2784507 + | CCGTTCTTA---------AACTAAAGTTTCCTTTTAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCGAA-AGCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGCGAACAATCAGC------------------GAAAACGGA |
| droSec1 | super_2:3244745-3244871 + | CCGTTCTTA---------AAATAAAGTTTCCTTTTAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCGAA-AGCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGTGAACAATCAGC------------------GAAAACGGA |
| droYak2 | chr3L:3795918-3796050 + | TCGTTCTATATCCGTTATAT---CAGTTTCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TACGAA-AGCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGTGAAAAATCAGC------------------GAAAACGGA |
| droEre2 | scaffold_4784:5933708-5933834 + | TCGTTCTAT---------CACCACAGTTTCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCAAA-AGCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGCGAACAAGCAGC------------------GAAAACGGA |
| droAna3 | scaffold_13337:7556585-7556720 + | CCGTTCTAA-TTCGAAA-ATCGAAAATTTCCTTATAGATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCGAA-AGTCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGCGACAAGACATC----------------ACAAAAACGGA |
| dp4 | chrXR_group8:3299974-3300083 - | ---------------------------TGCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTTGAAAACCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGAAGAAATGCATC------------------AAAAGCGGA |
| droPer1 | super_65:103931-104040 - | ---------------------------TGCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTTGAAAACCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGAAGAAATGCATC------------------AAAAGCGGA |
| droWil1 | scaffold_181136:1999668-1999787 + | TTTTCCTTTAT--GTAA-TATTAATATATCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTTGGA------TTCAGACATAGCCTATGAGAGGTTAGGTGTACCAAGGAGAAAATACAAC------------------A-------- |
| droVir3 | scaffold_12758:910696-910814 - | GCGTCCGAC--------------AAATTTCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCGAT-ATACGGTCAGACATAGCCTATGAGAGGTTAGGTGTACCAAGGGG-ACAAGCAGC------------------GAGGGCG-- |
| droMoj3 | scaffold_6680:1963284-1963406 - | ATTTTTTTT-TGCG------GT-GCATTTCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTTGAT-ATAAGATCAGACATAGCCTATGAGAGGTTAGGTGTAGCAAGGGG-TCAAACAGC------------------AGA----GA |
| droGri2 | scaffold_15110:607073-607202 + | ---------------------------TTCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCACTTTTTCA-ATTCAATCAGACATAGCCTATGAGAGGTTAGGTGTACAAATGGATCAAAGCAACAAGGCTGCTGTCGGGGACTAAAAATGA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:3251007-3251133 + | CCGTTCTTT---------AACTATAGTTTCCTTCTAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCGAA-AGCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGCGAACAATCAGC------------------GAAAACGGA |
| droSim1 | chr3L:2784381-2784507 + | CCGTTCTTA---------AACTAAAGTTTCCTTTTAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCGAA-AGCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGCGAACAATCAGC------------------GAAAACGGA |
| droSec1 | super_2:3244745-3244871 + | CCGTTCTTA---------AAATAAAGTTTCCTTTTAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCGAA-AGCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGTGAACAATCAGC------------------GAAAACGGA |
| droYak2 | chr3L:3795918-3796050 + | TCGTTCTATATCCGTTATAT---CAGTTTCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TACGAA-AGCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGTGAAAAATCAGC------------------GAAAACGGA |
| droEre2 | scaffold_4784:5933708-5933834 + | TCGTTCTAT---------CACCACAGTTTCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCAAA-AGCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGCGAACAAGCAGC------------------GAAAACGGA |
| droAna3 | scaffold_13337:7556585-7556720 + | CCGTTCTAA-TTCGAAA-ATCGAAAATTTCCTTATAGATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCGAA-AGTCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGCGACAAGACATC----------------ACAAAAACGGA |
| dp4 | chrXR_group8:3299974-3300083 - | ---------------------------TGCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTTGAAAACCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGAAGAAATGCATC------------------AAAAGCGGA |
| droPer1 | super_65:103931-104040 - | ---------------------------TGCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTTGAAAACCCGATCAGACATAGCCTATAAGAGGTTAGGTGTACCAAGGAAGAAATGCATC------------------AAAAGCGGA |
| droWil1 | scaffold_181136:1999668-1999787 + | TTTTCCTTTAT--GTAA-TATTAATATATCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTTGGA------TTCAGACATAGCCTATGAGAGGTTAGGTGTACCAAGGAGAAAATACAAC------------------A-------- |
| droVir3 | scaffold_12758:910696-910814 - | GCGTCCGAC--------------AAATTTCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTCGAT-ATACGGTCAGACATAGCCTATGAGAGGTTAGGTGTACCAAGGGG-ACAAGCAGC------------------GAGGGCG-- |
| droMoj3 | scaffold_6680:1963284-1963406 - | ATTTTTTTT-TGCG------GT-GCATTTCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCA--TTTGAT-ATAAGATCAGACATAGCCTATGAGAGGTTAGGTGTAGCAAGGGG-TCAAACAGC------------------AGA----GA |
| droGri2 | scaffold_15110:607073-607202 + | ---------------------------TTCCTTATAAATCTAGCCTCTACTAGGCTTTGTCTGTGCACTTTTTCA-ATTCAATCAGACATAGCCTATGAGAGGTTAGGTGTACAAATGGATCAAAGCAACAAGGCTGCTGTCGGGGACTAAAAATGA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-34.2, p-value=0.009901 | dG=-34.1, p-value=0.009901 |
|---|---|
 |
 |
| dG=-34.2, p-value=0.009901 | dG=-34.1, p-value=0.009901 |
|---|---|
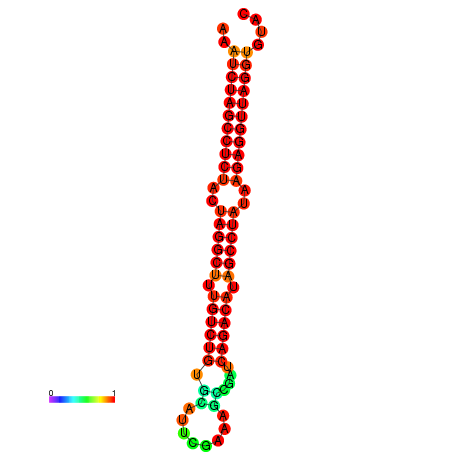 |
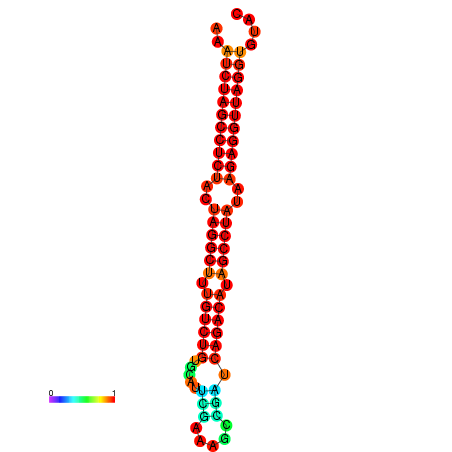 |
| dG=-34.2, p-value=0.009901 | dG=-34.1, p-value=0.009901 |
|---|---|
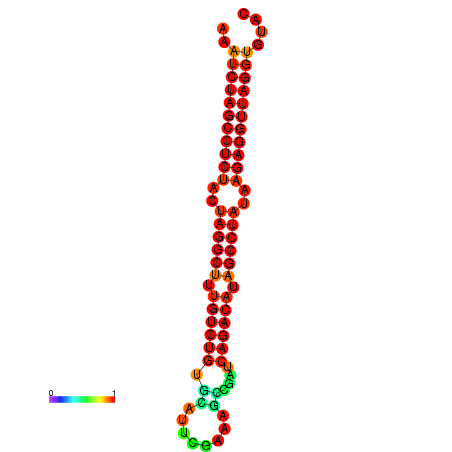 |
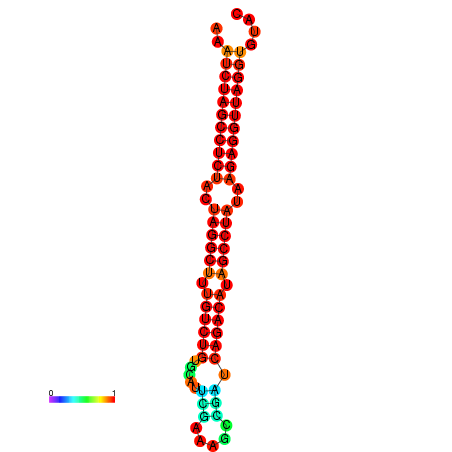 |
| dG=-34.2, p-value=0.009901 |
|---|
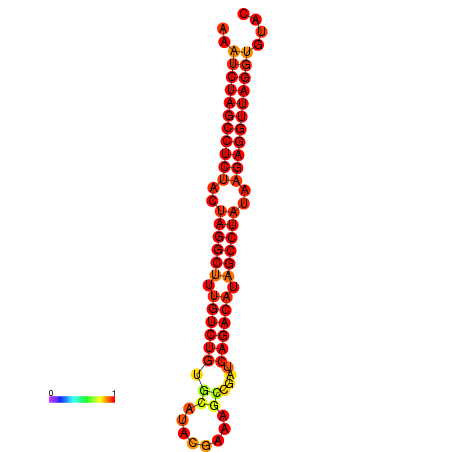 |
| dG=-34.2, p-value=0.009901 |
|---|
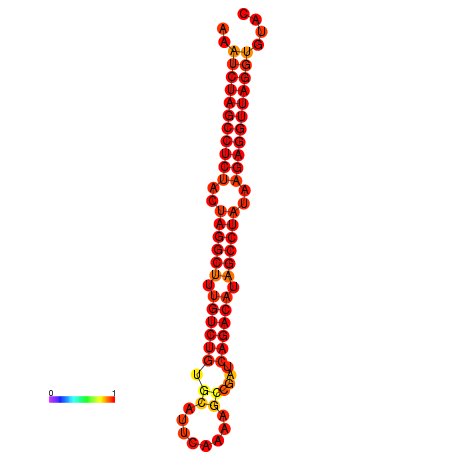 |
| dG=-34.8, p-value=0.009901 | dG=-34.1, p-value=0.009901 |
|---|---|
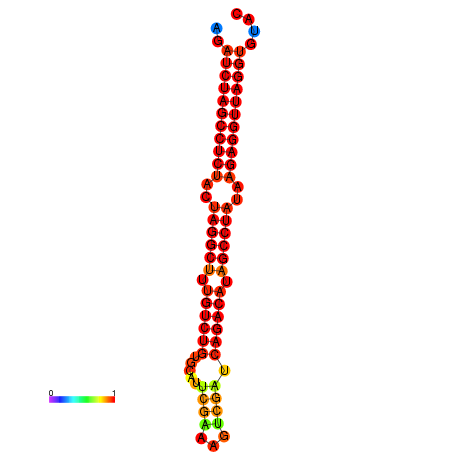 |
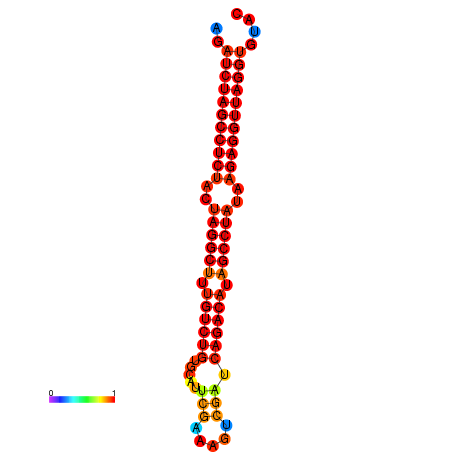 |
| dG=-33.1, p-value=0.009901 | dG=-32.3, p-value=0.009901 |
|---|---|
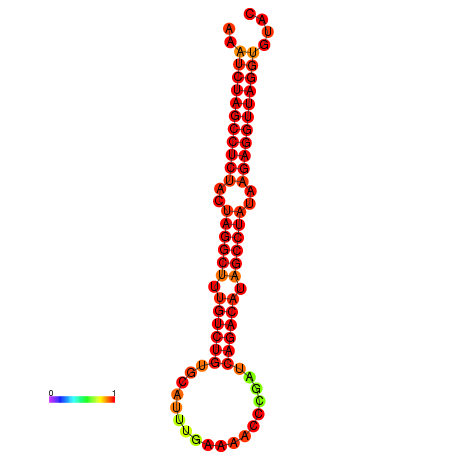 |
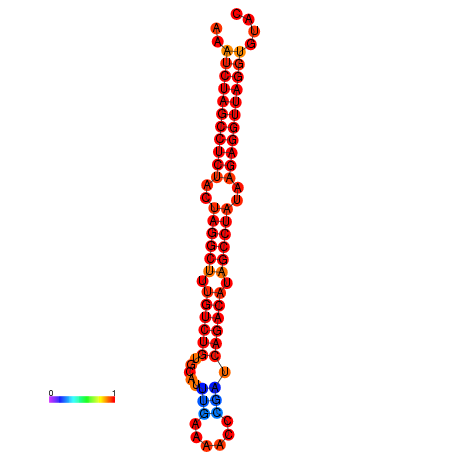 |
| dG=-33.1, p-value=0.009901 | dG=-32.3, p-value=0.009901 |
|---|---|
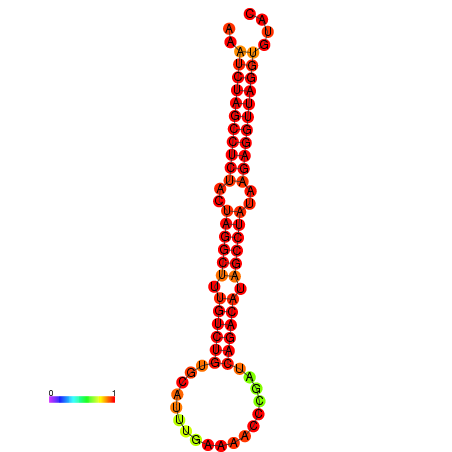 |
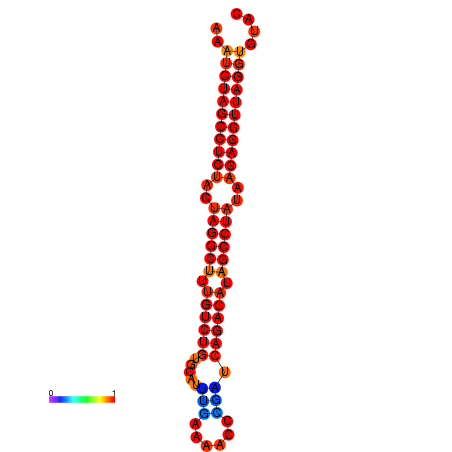 |
| dG=-33.9, p-value=0.009901 |
|---|
 |
| dG=-33.9, p-value=0.009901 | dG=-33.8, p-value=0.009901 | dG=-33.5, p-value=0.009901 | dG=-33.2, p-value=0.009901 |
|---|---|---|---|
 |
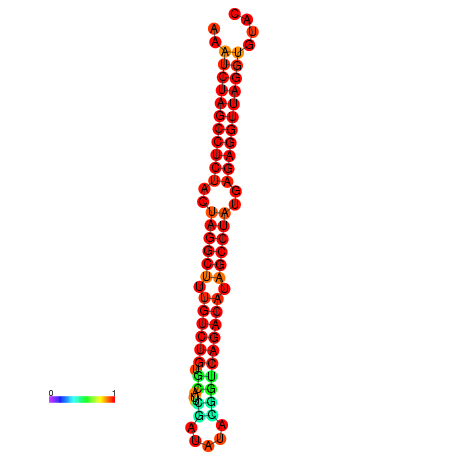 |
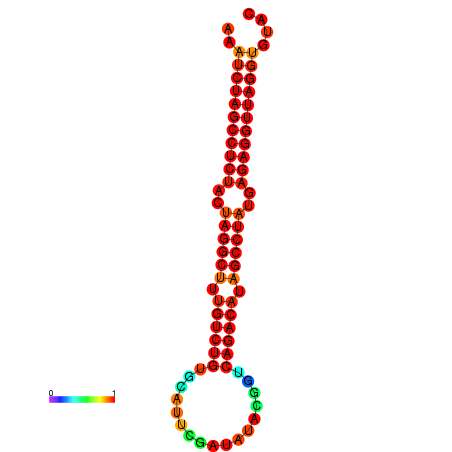 |
 |
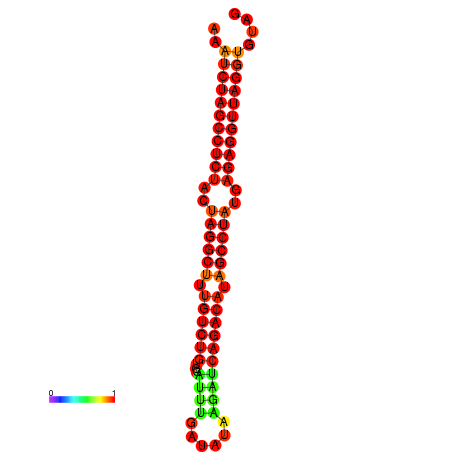
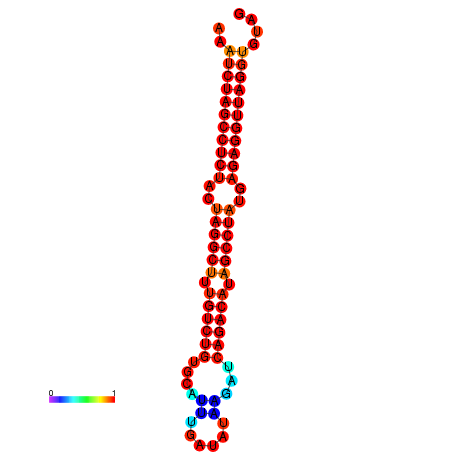
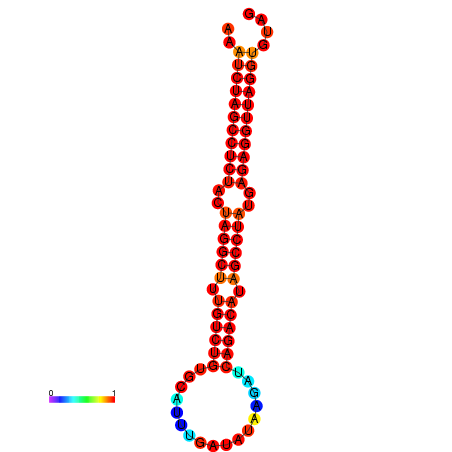
| dG=-34.4, p-value=0.009901 | dG=-33.6, p-value=0.009901 | dG=-33.5, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-33.4, p-value=0.009901 |
|---|
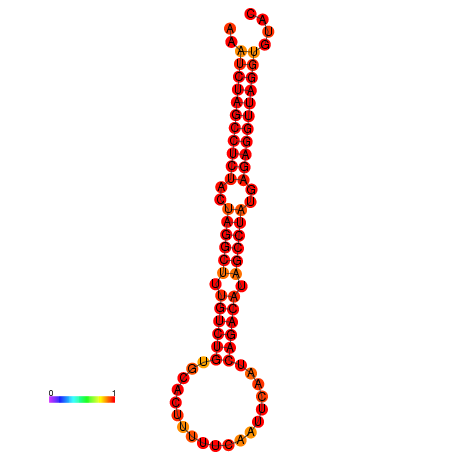 |
Generated: 03/07/2013 at 05:03 PM