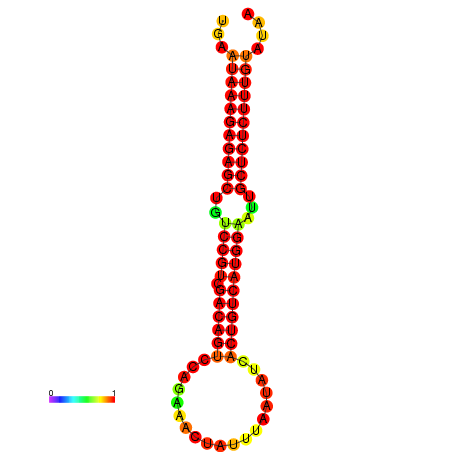
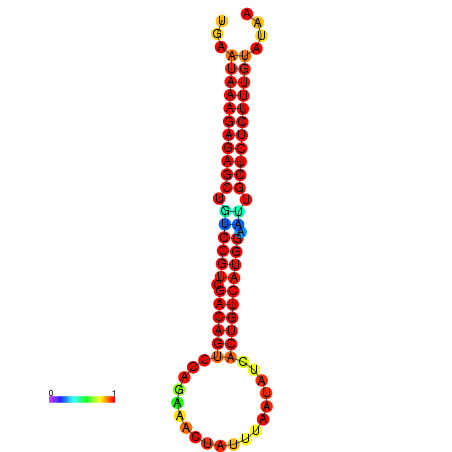
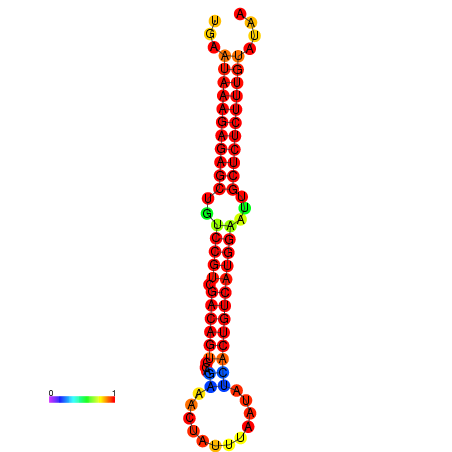
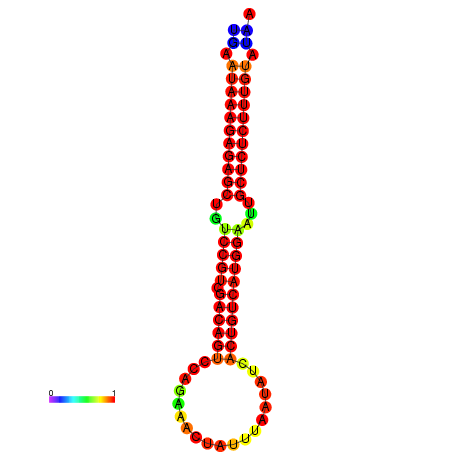
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:8057863-8057982 - | AATCGAATC-CAAATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--AACTA-TT-TAATATCACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGATGCGCTGTGAG |
| droSim1 | chr2R_random:2148092-2148211 - | AATCGAATC-CAAATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--AAGCA-TT-TAATATCACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGATGCGCTGTGAG |
| droSec1 | super_1:5602965-5603084 - | AATCGAATC-CAAATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--AACCA-TT-TAATATCACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGATGCGCTGTGAT |
| droYak2 | chr2R:8893333-8893452 + | AATCGAATC-CATATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--ATTCA-TT-TAATATCACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGATGCGCTATGAG |
| droEre2 | scaffold_4845:17736182-17736301 + | AATCGAATA-CATATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--AACCA-TT-TAATATCACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGATGCACTATGAG |
| droAna3 | scaffold_13266:2568740-2568853 + | CT---ATCT-CGAATGCGAATACGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--TACCT---ATAAC---ACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGCTTCGCTCCGAT |
| dp4 | chr3:418413-418523 - | AA---AACA-CGAATGCGAATAGGTGAATAAAGAGAGCTGTCCGTCGACAGTCAACG--TACCAATT-TAAT---ACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGTTGCGC----AG |
| droPer1 | super_2:605310-605420 - | AA---AACA-CGAATGCGAATAGGTGAATAAAGAGAGCTGTCCGTCGACAGTCAACG--TACCAATT-TAAT---ACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGTTGCGC----AG |
| droWil1 | scaffold_181141:3241875-3241979 + | AG---A-----AAAAGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGTTAAACGA-TTTTAAA---ACTGTCATGGAATTGCTCTCTTTGTATGATATTCGACGT--------GAC |
| droVir3 | scaffold_12875:19829913-19830024 + | AATCGTACC-CAAAAGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGT--AACAA-ATGTAAT---ACTGTCATGGAATTGCTCTCTTTGTATGATATTCGACGTTGCCC------ |
| droMoj3 | scaffold_6496:6376225-6376337 - | AATCGTAGCGAAAATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGT--AAAGA-TTGTAAA---ACTGTCATGGAATTGCTCTCTTTGTATGATATTCGACGTTGCCC------ |
| droGri2 | scaffold_15245:18178188-18178294 + | AATT---AT-GAAAAGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGT--ATAAA-TTTTAAT---ACTGTCATGGAATTGCTCTCTTTGTATGATATTCGACGTTGC-------- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:8057863-8057982 - | AATCGAATC-CAAATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--AACTA-TT-TAATATCACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGATGCGCTGTGAG |
| droSim1 | chr2R_random:2148092-2148211 - | AATCGAATC-CAAATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--AAGCA-TT-TAATATCACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGATGCGCTGTGAG |
| droSec1 | super_1:5602965-5603084 - | AATCGAATC-CAAATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--AACCA-TT-TAATATCACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGATGCGCTGTGAT |
| droYak2 | chr2R:8893333-8893452 + | AATCGAATC-CATATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--ATTCA-TT-TAATATCACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGATGCGCTATGAG |
| droEre2 | scaffold_4845:17736182-17736301 + | AATCGAATA-CATATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--AACCA-TT-TAATATCACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGATGCACTATGAG |
| droAna3 | scaffold_13266:2568740-2568853 + | CT---ATCT-CGAATGCGAATACGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGA--TACCT---ATAAC---ACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGCTTCGCTCCGAT |
| dp4 | chr3:418413-418523 - | AA---AACA-CGAATGCGAATAGGTGAATAAAGAGAGCTGTCCGTCGACAGTCAACG--TACCAATT-TAAT---ACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGTTGCGC----AG |
| droPer1 | super_2:605310-605420 - | AA---AACA-CGAATGCGAATAGGTGAATAAAGAGAGCTGTCCGTCGACAGTCAACG--TACCAATT-TAAT---ACTGTCATGGAATTGCTCTCTTTGTATAATATTCGACGTTGCGC----AG |
| droWil1 | scaffold_181141:3241875-3241979 + | AG---A-----AAAAGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGTTAAACGA-TTTTAAA---ACTGTCATGGAATTGCTCTCTTTGTATGATATTCGACGT--------GAC |
| droVir3 | scaffold_12875:19829913-19830024 + | AATCGTACC-CAAAAGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGT--AACAA-ATGTAAT---ACTGTCATGGAATTGCTCTCTTTGTATGATATTCGACGTTGCCC------ |
| droMoj3 | scaffold_6496:6376225-6376337 - | AATCGTAGCGAAAATGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGT--AAAGA-TTGTAAA---ACTGTCATGGAATTGCTCTCTTTGTATGATATTCGACGTTGCCC------ |
| droGri2 | scaffold_15245:18178188-18178294 + | AATT---AT-GAAAAGCGAATAAGTGAATAAAGAGAGCTGTCCGTCGACAGTCCAGT--ATAAA-TTTTAAT---ACTGTCATGGAATTGCTCTCTTTGTATGATATTCGACGTTGC-------- |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-26.7, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
| dG=-26.7, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|
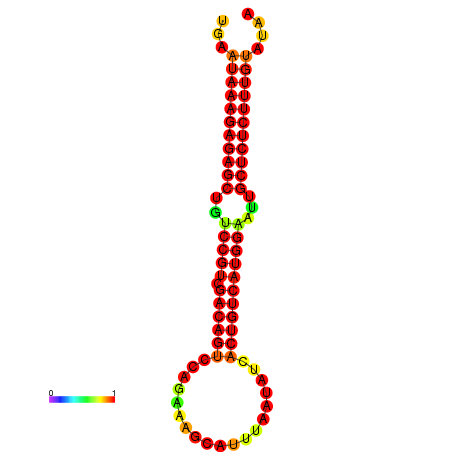 |
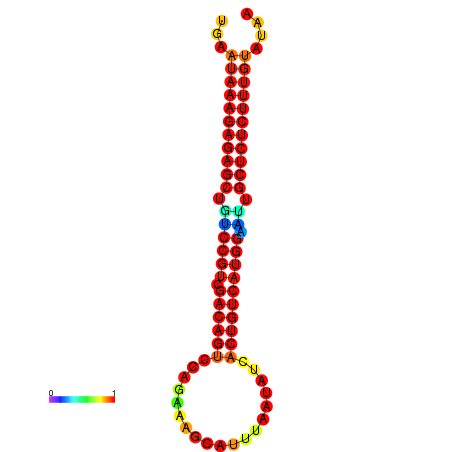 |
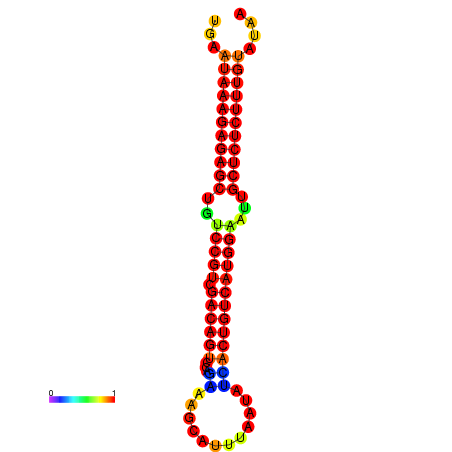 |
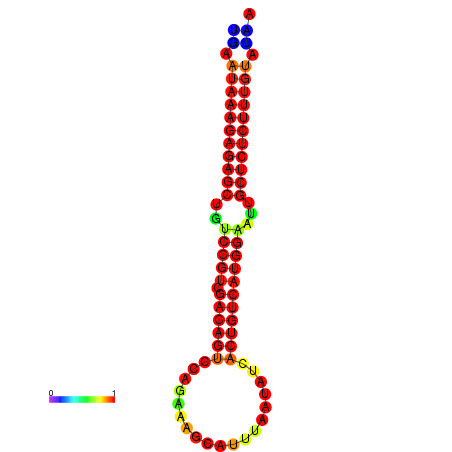 |
| dG=-26.7, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|
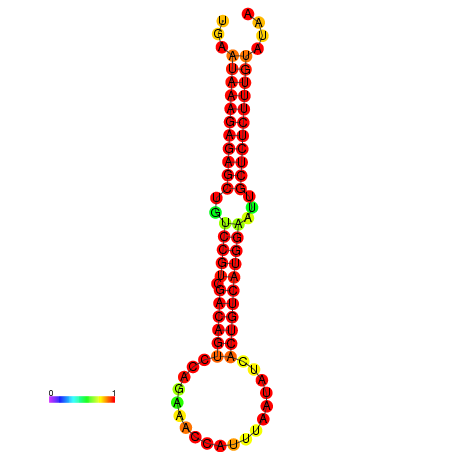 |
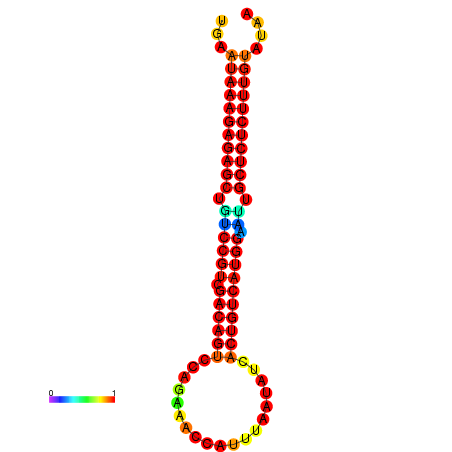 |
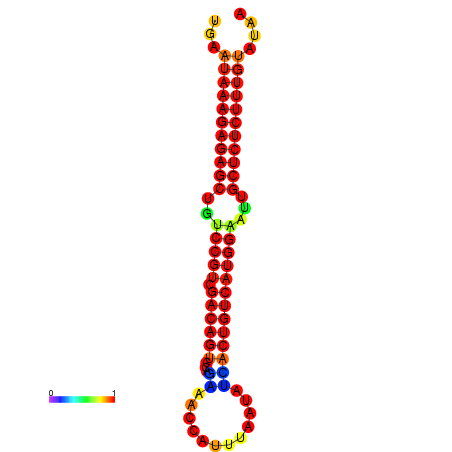 |
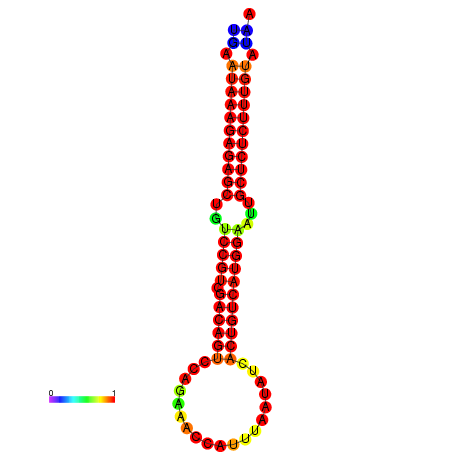 |
| dG=-26.7, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-26.7, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
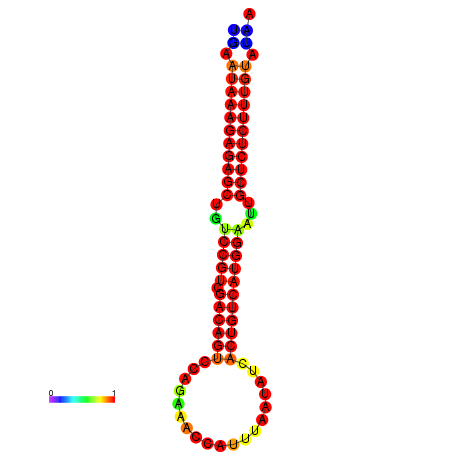 |
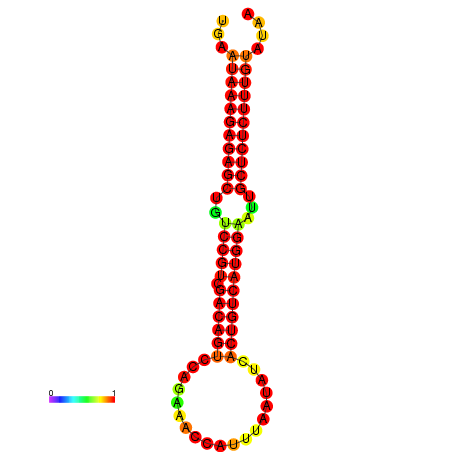
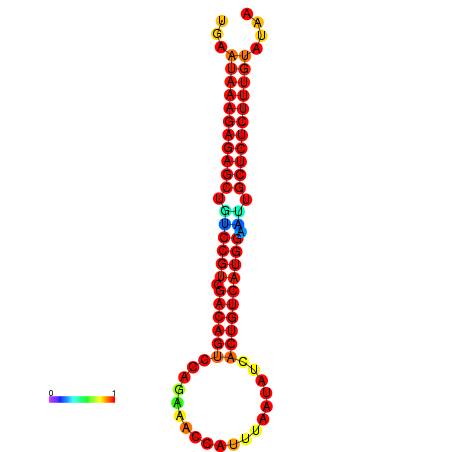
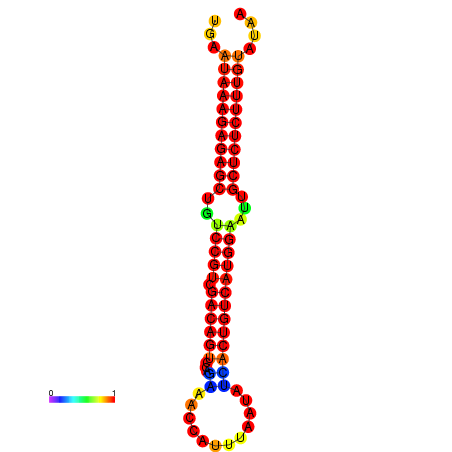
| dG=-27.0, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-26.0, p-value=0.009901 |
|---|---|---|---|---|
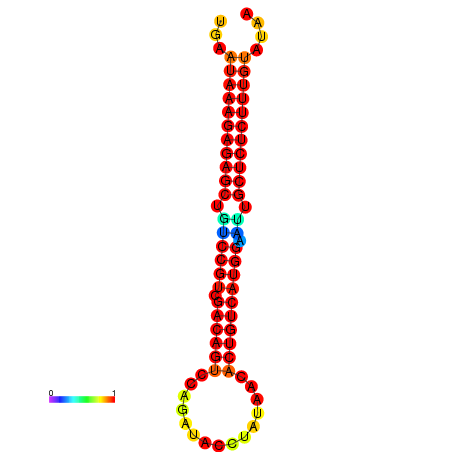
 |
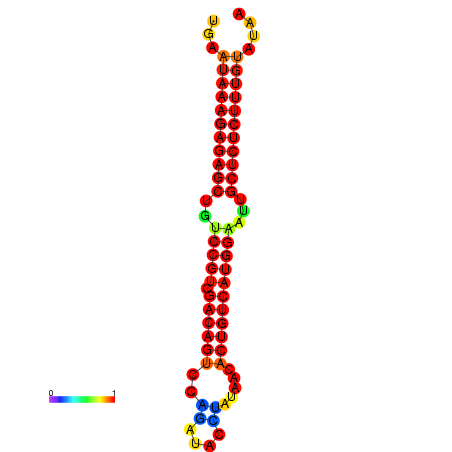
 |
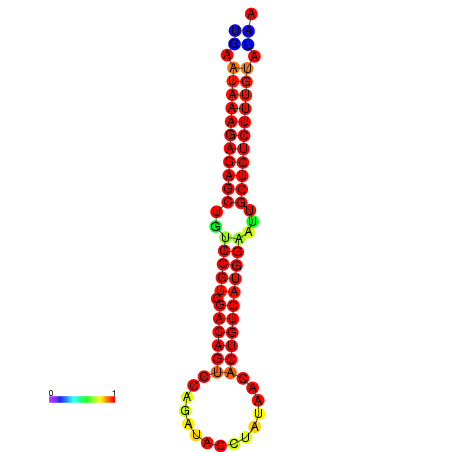
 |
 |
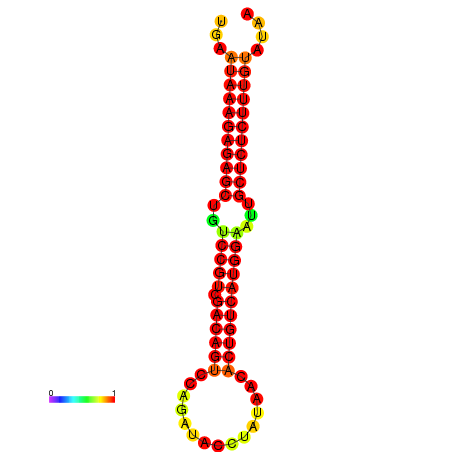
 |
| dG=-26.7, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|
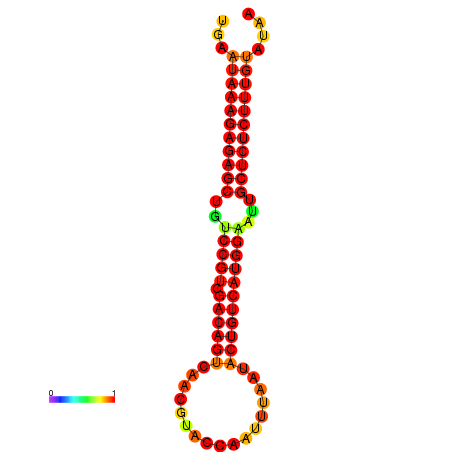 |
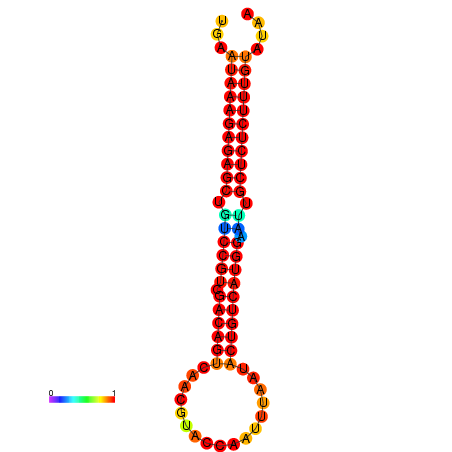 |
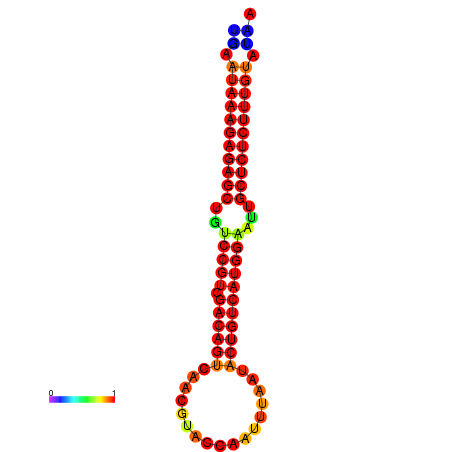 |
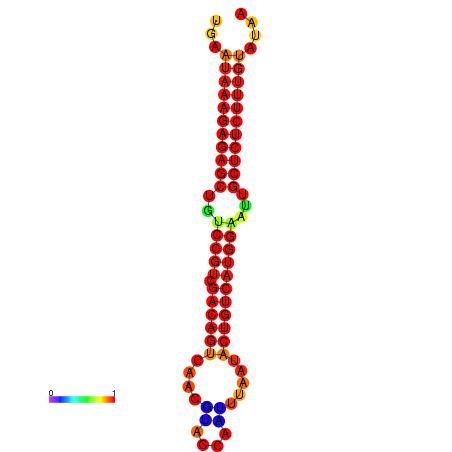 |
| dG=-26.7, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|
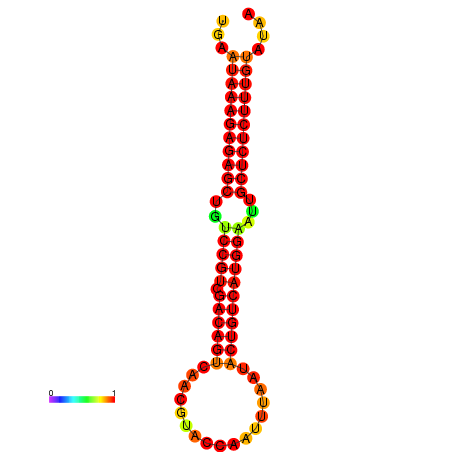 |
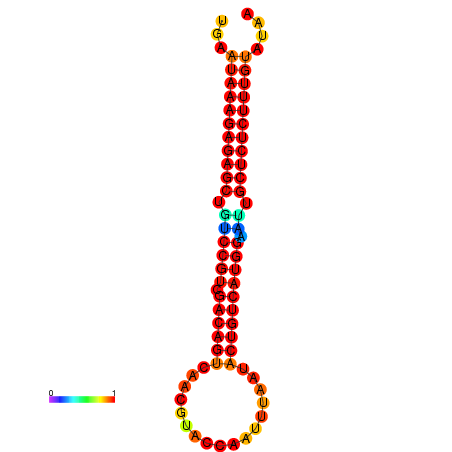 |
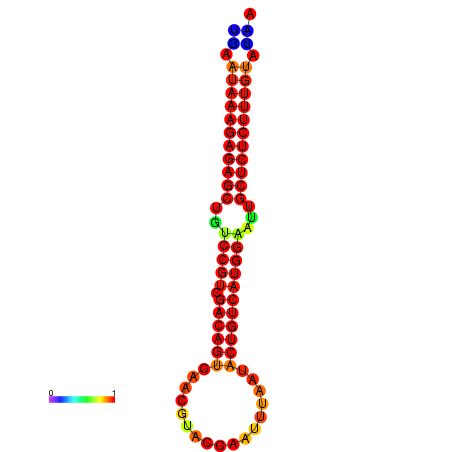 |
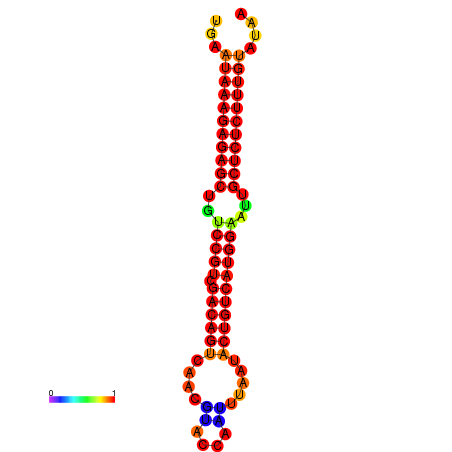 |
| dG=-27.2, p-value=0.009901 | dG=-26.8, p-value=0.009901 | dG=-26.8, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.5, p-value=0.009901 |
|---|---|---|---|---|
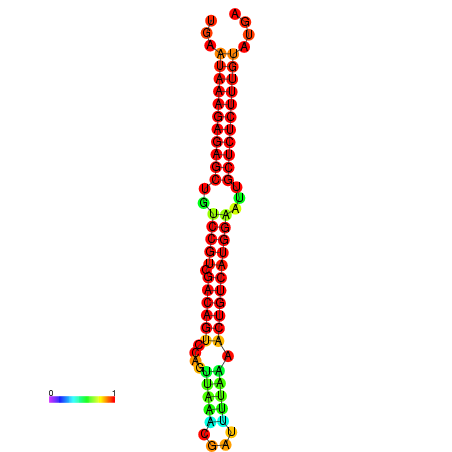 |
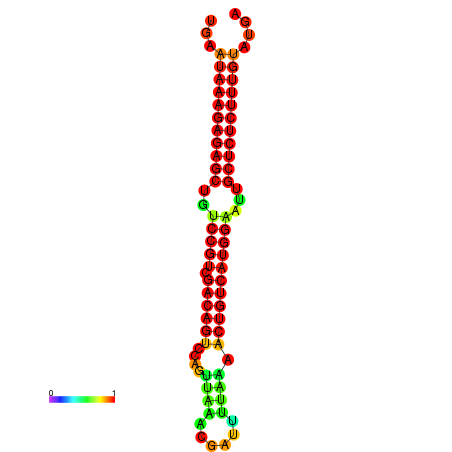 |
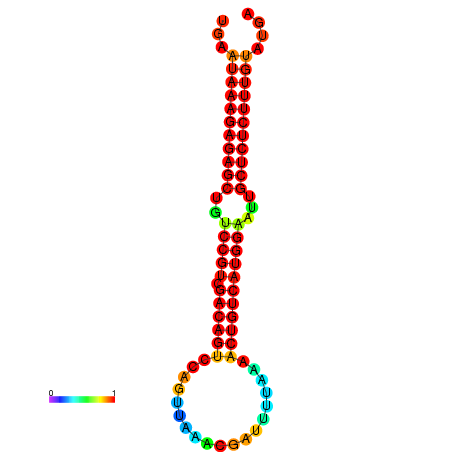 |
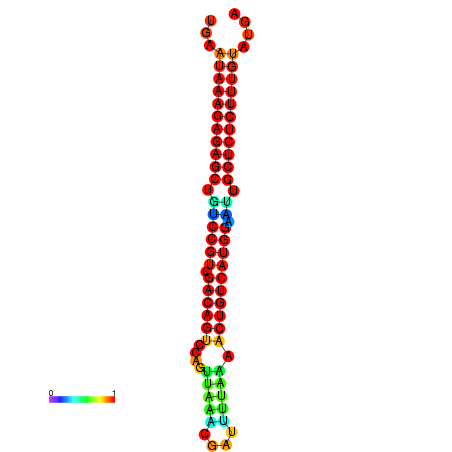 |
 |
| dG=-27.2, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
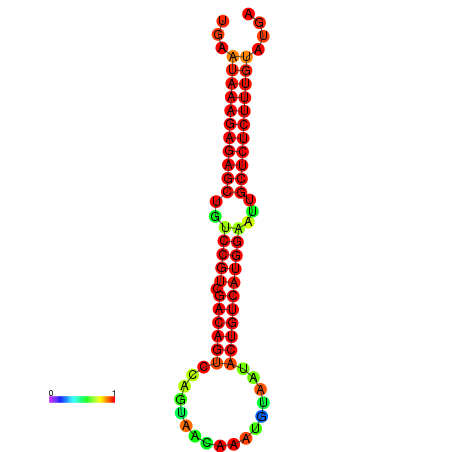 |
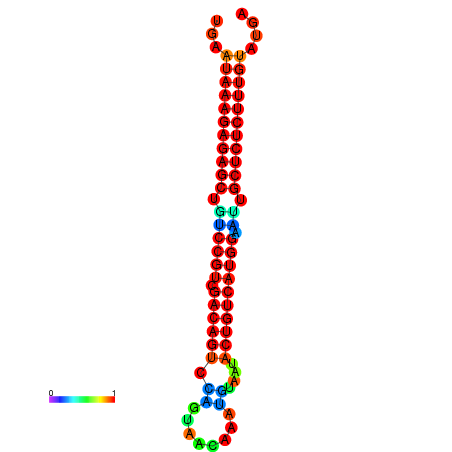 |
| dG=-28.3, p-value=0.009901 | dG=-27.8, p-value=0.009901 |
|---|---|
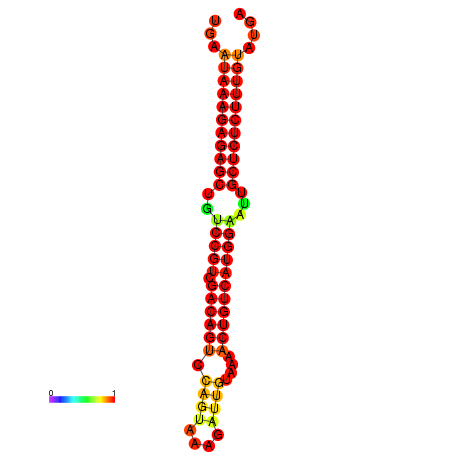 |
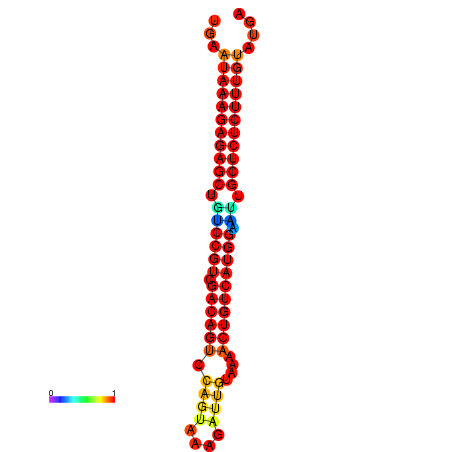 |
| dG=-26.7, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-25.9, p-value=0.009901 |
|---|---|---|---|---|
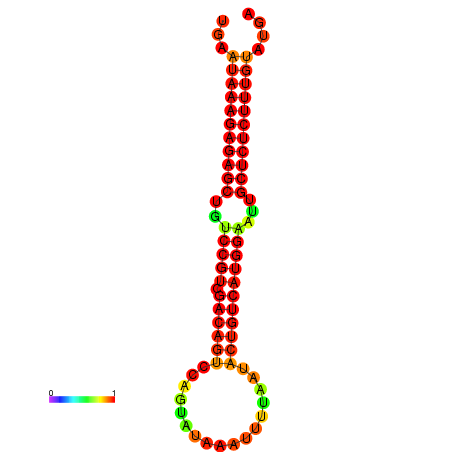 |
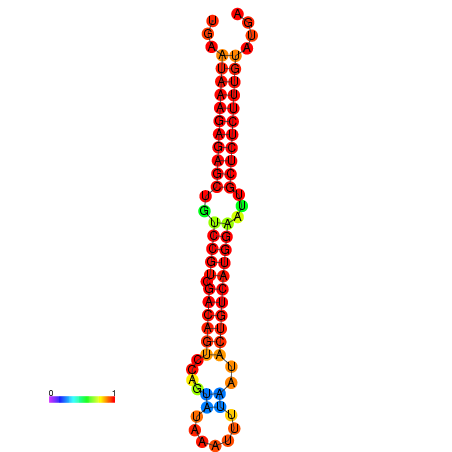 |
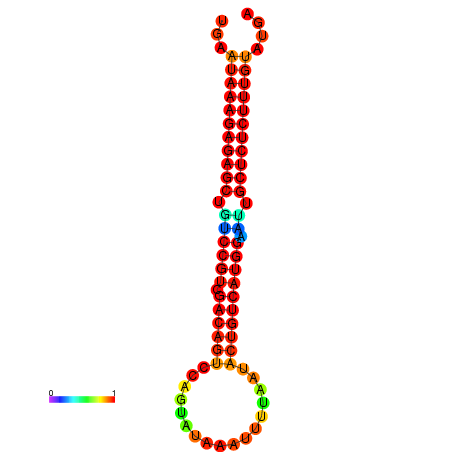 |
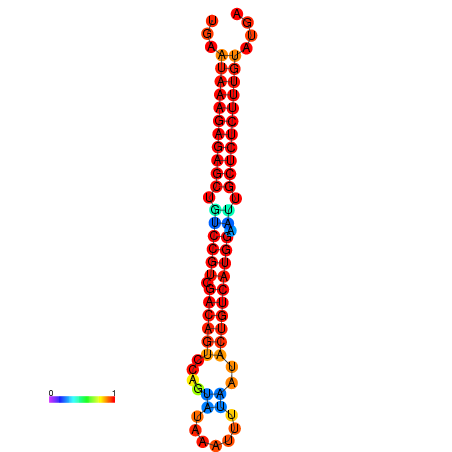 |
 |
Generated: 03/07/2013 at 05:03 PM