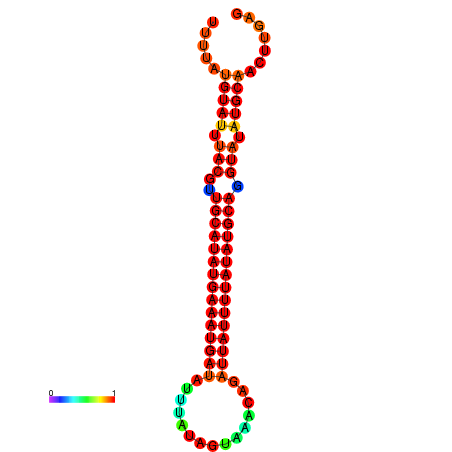
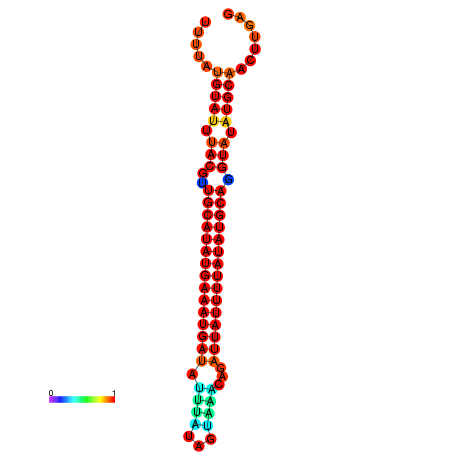
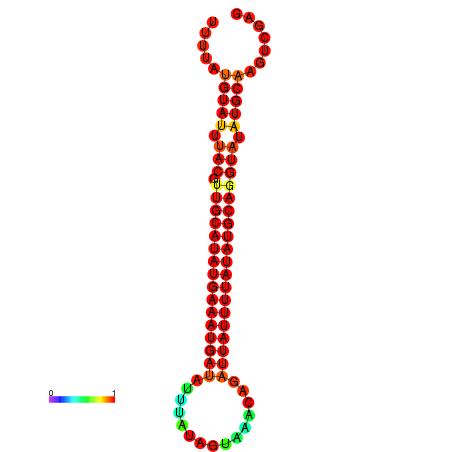
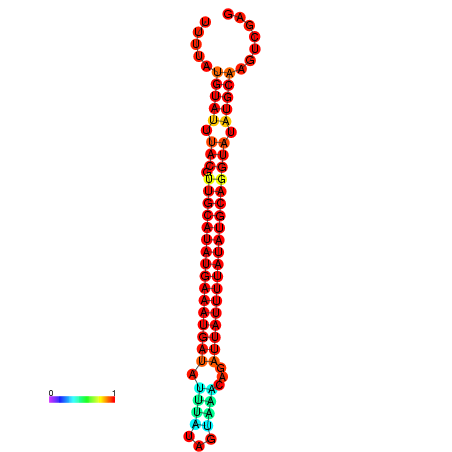
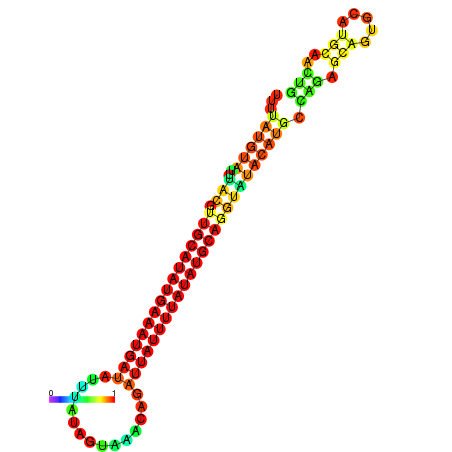
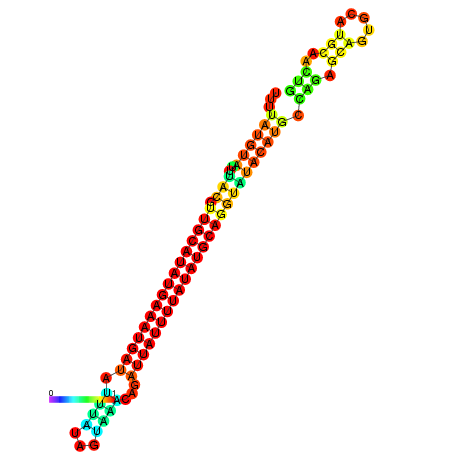
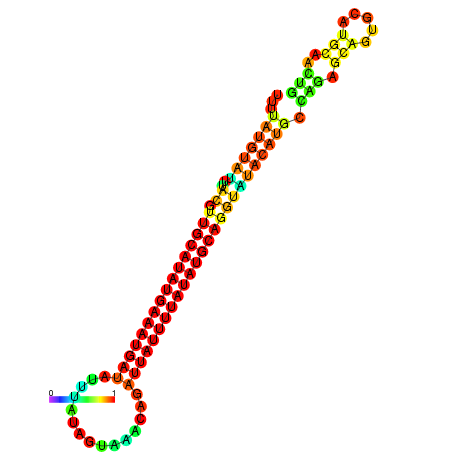
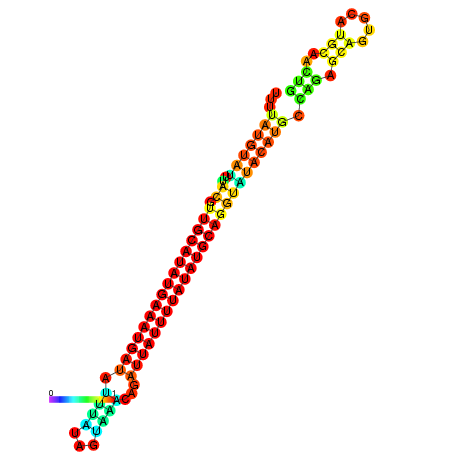
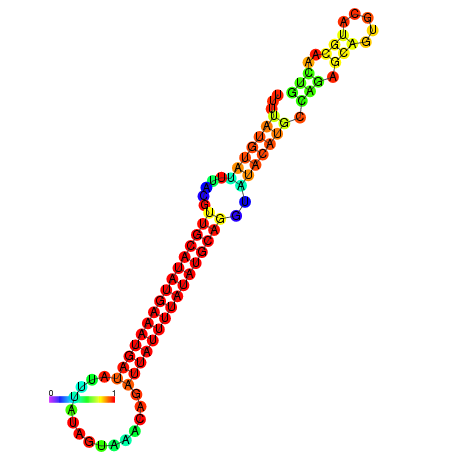
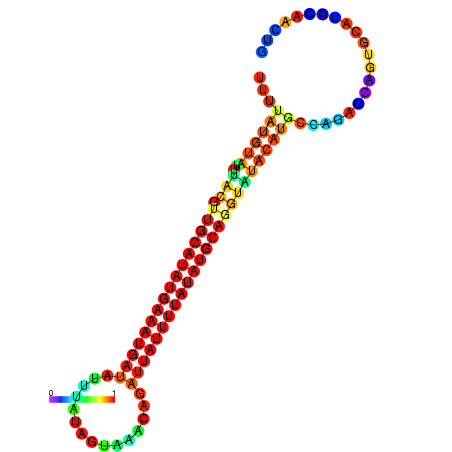
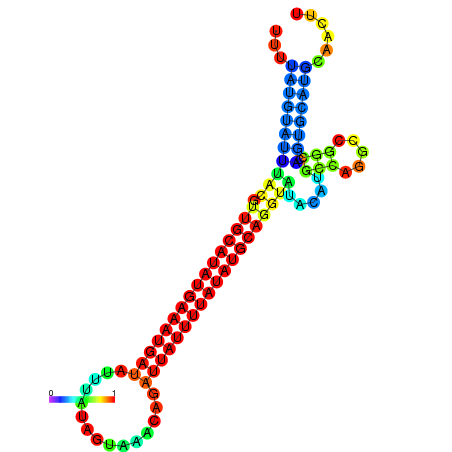
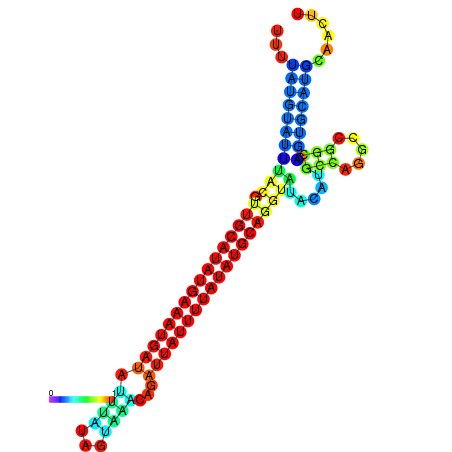
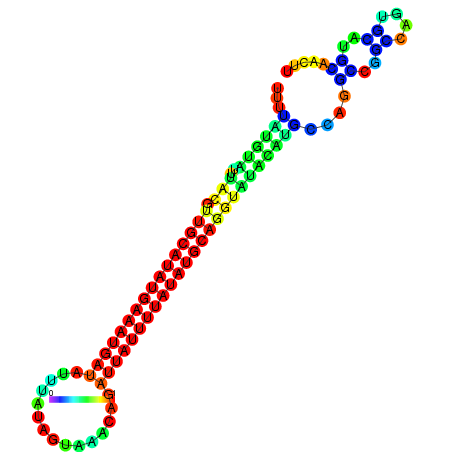
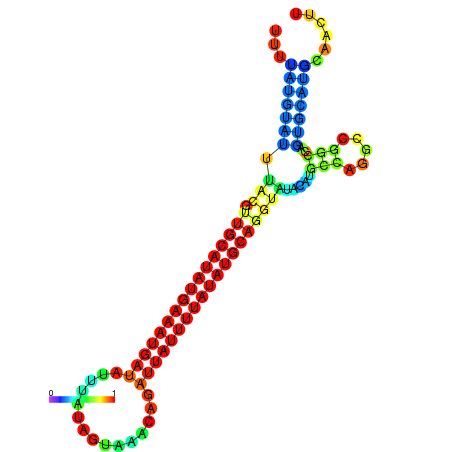
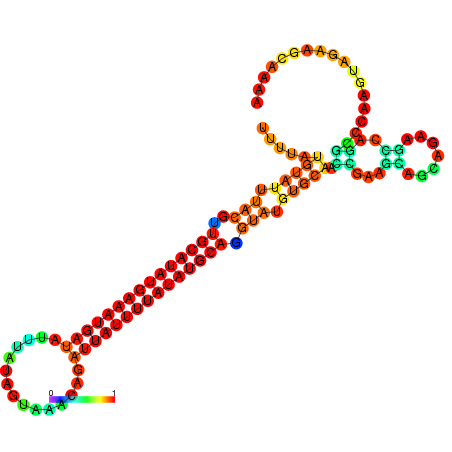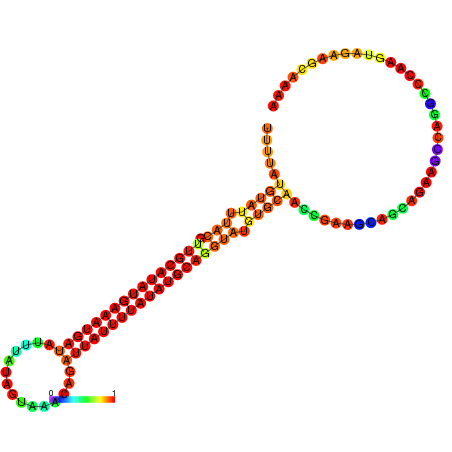| dm3 |
chr2R:4185596-4185720 + |
CTGACGT-------------------G--------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTATGCTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATA--TGCAA-------------------------------------GTCGA------GGTCCTCCA-----------CACT-------GCACTCGC-CGC-CTCGA |
| droSim1 |
zee54c10.b1:599-724 + |
CTGACGT-------------------G--------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTGTGCTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATA--TGCAA-------------------------------------GTCGA------GGTCCTGCA-----------CACT-------GCACTCGC-CGC-CTCGA |
| droSec1 |
super_1:1832273-1832397 + |
CTGACGT-------------------G--------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTGTGCTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATA--TGCAA-------------------------------------GTCGA------GGTCCTGCA-----------CACT-------GCACTCGC-CGC-CTCGA |
| droYak2 |
chr2L:16840791-16840915 + |
CTGACGT-------------------G--------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTGTGCTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATA--TGCAA-------------------------------------CTTGA------GGTCCTGCA-----------CACT-------GCACTCTC-CCG-CTCGA |
| droEre2 |
scaffold_4929:18446687-18446817 - |
CTGACGT-------------------G--------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTGTGCTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATA--TGCAA-------------------------------------GTCGA------GGTCCAGCA-----------CACTGCACACTGCACTTGC-C-C-CTCGA |
| droAna3 |
scaffold_13266:1806990-1807233 - |
TTGAAAT-------------------G-----TGTGTATGTGGG--------TG----------GGTG----GGTGTGTGTGCACTTTCTGCCATCCAGACTGCCAGCCAGCCATCTAGCCAGCCTGCCTGACTGCTTTCCTTGCCACTT-----GTGTGTATGTGTGTGTG-----------------TGTATGAGCTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATA--TGCAA-------------------------------------G-------------CCTGAACAACTTTGGAACACT-------GCAATCGC-CTT-CTGGA |
| dp4 |
chr3:2170016-2170201 + |
TTGATGC-------------------C-----tgtgtgtgtgtg--------------------------tgtgtgtgtgtCCAGTCGGTGTCATGA-GG-------------------------------------------------CGTATGCC----GTGTGTGTGTG-----------------TGTGTGAGACGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATG--TGCAG-------------------------------------CTGAA------GAGCCTGTC-----------CATT-------GCACAATC-C-C-CACCC |
| droPer1 |
super_2:2339680-2339867 + |
TTGATGC-------------------C-----TGTGTGTGTGTG----------------------------TGTGTGCGTCCAGTCGGTGTCATGA-GG-------------------------------------------------CGTATGCCgtgtgtgtgtgtgtg-----------------tgtgtgAGACGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATG--TGCAG-------------------------------------CTGAA------GAGCCTGTC-----------CATT-------GCACAATC-C-C-CTCCC |
| droWil1 |
scaffold_180701:589914-590141 + |
TTGATGCCCCAACACACACCCACCTCC-----TATCTATCTATCTATCTCTGTCTCACCATAACTGAg------------------------------------------------------------------------------------------tgtgtgtgtgtgtg-----------------tgtgtatgtTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATG--TGCAACCGAAGCAGCAGAAGCCAGGCC--CAA----------GTAGAAGCAAAAGGCCTGAA-----------CATT-------GCACTTGGGC-CATACGC |
| droVir3 |
scaffold_12875:9206154-9206327 + |
TTGATGT-------------------CGTCTTTGTGTG-----CCAT------------------GTGTGTGTGTGTGTGTCCATT-------------------------------------------------------------------------------------T-----------------TGTGTGTGTTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATACATGCCA------------------GA----GCAGTGCATGCAACTG------------C-CCA-----------CAAT-------GCACATTT-C-TATCCGC |
| droMoj3 |
scaffold_6496:17934933-17935113 - |
TTGATGT-------------------CGTCGTTGTGTA-----CCATGCATTCG----------TGTG------------------------------------------------------------------------------------------TGCCTGTGTGTGTG-----------------CGTGTGTGTTAGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATACATGCCA------------------GGCCGGCCAGTGCATGCAACTT------------C-TCT-----------CAAT-------GCATATTC-C-AATCCAC |
| droGri2 |
scaffold_15245:2520625-2520797 + |
TTGATGT-------------------TGTCGTTGAGT------CTAtgtctgtg----------tgtg------------------------------------------------------------------------------------------tgtgtgtgtgtgtgtgtgcgagtgtCCAGTTCGTGTGTGTTGGCTTTTATGTATTTACGTTGCATATGAAATGATATTTATAGTAAACAGATTATTTTATATGCAGGTATACATGCAA-------------------------------------CTC------------CTTCT-----------CAAT-------GGATCCCC--------AC |