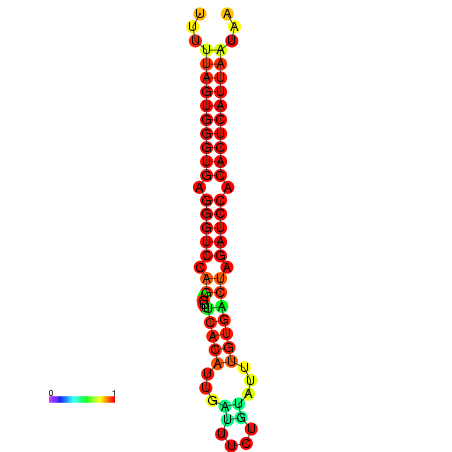
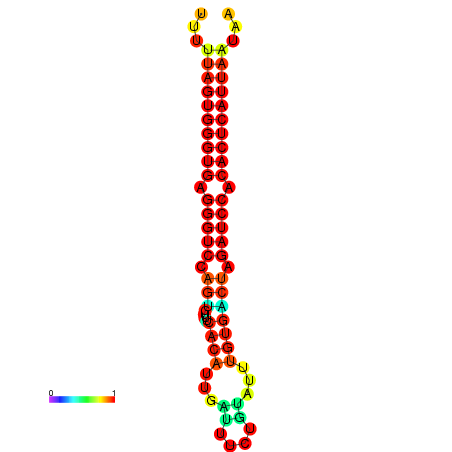
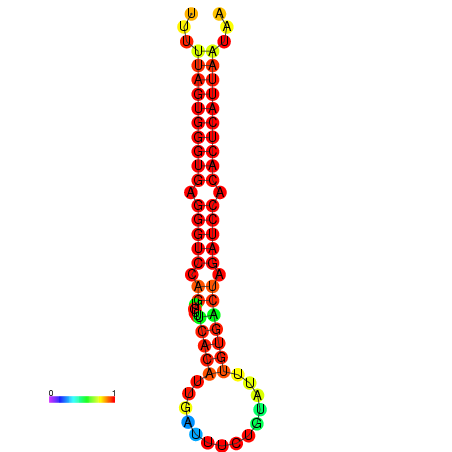
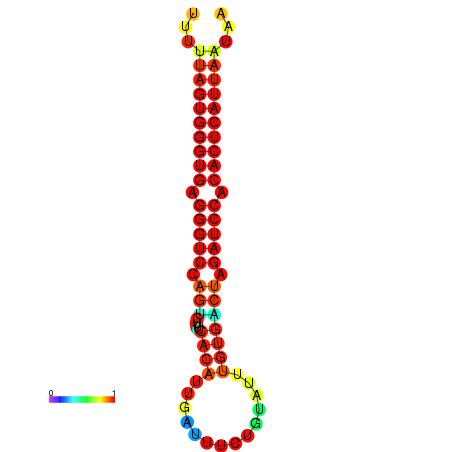
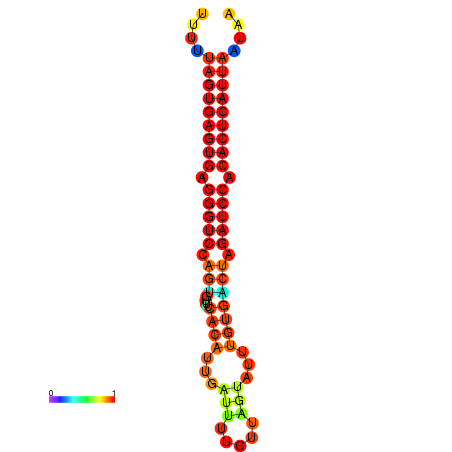
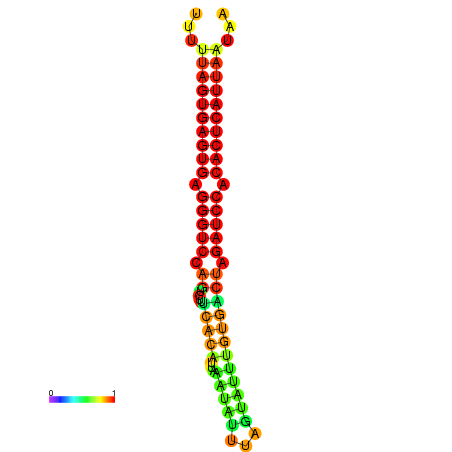
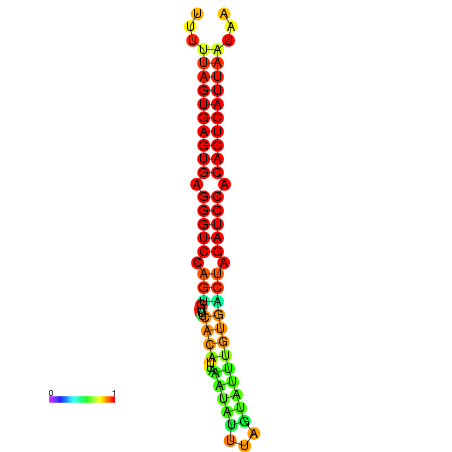
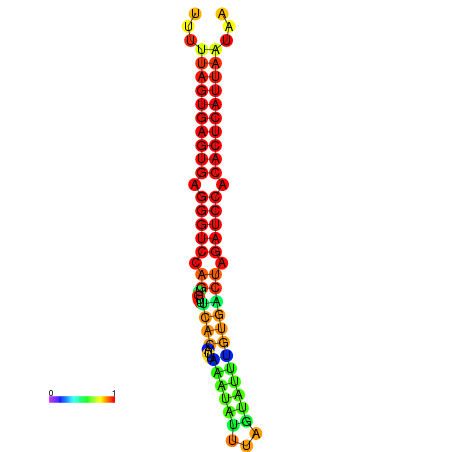
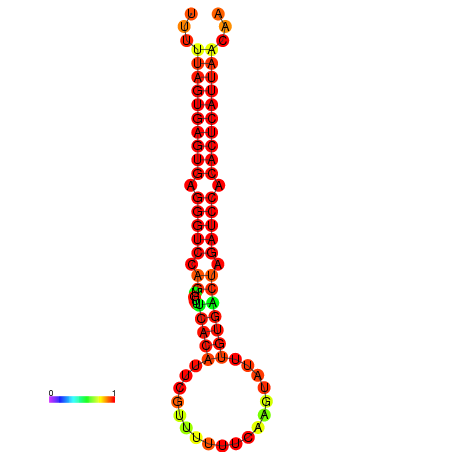
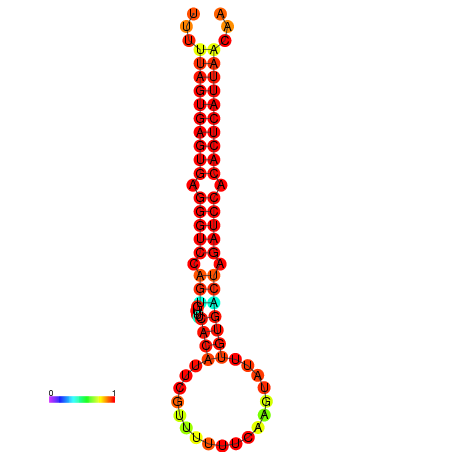
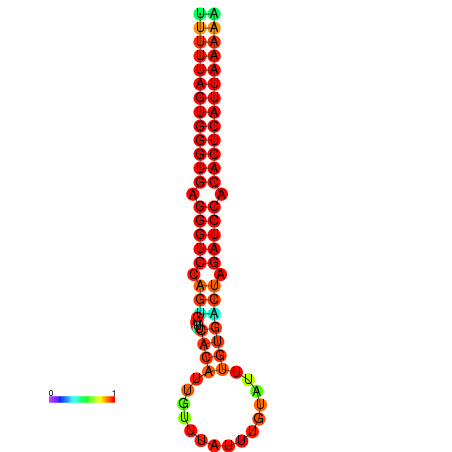
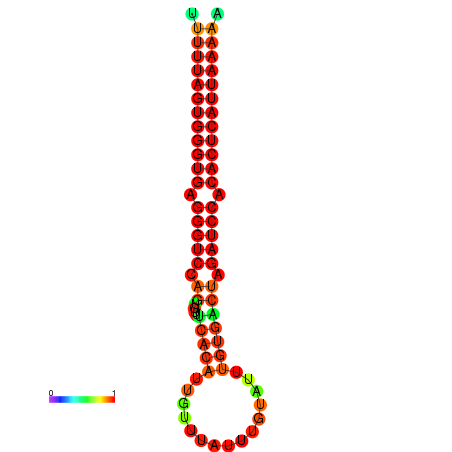
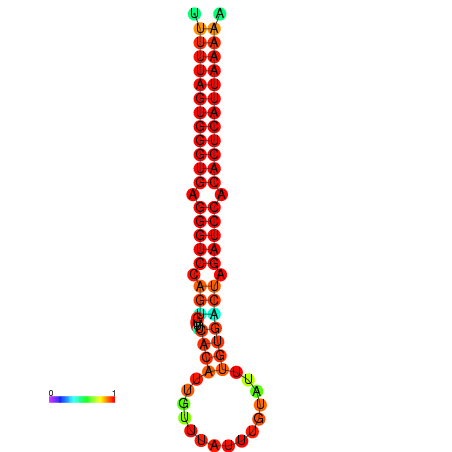
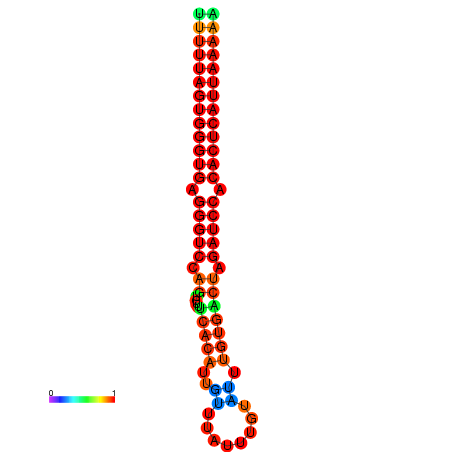
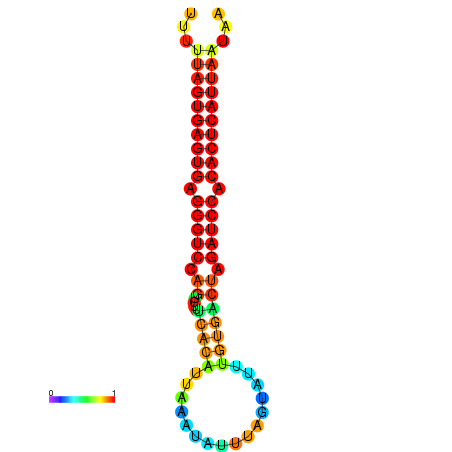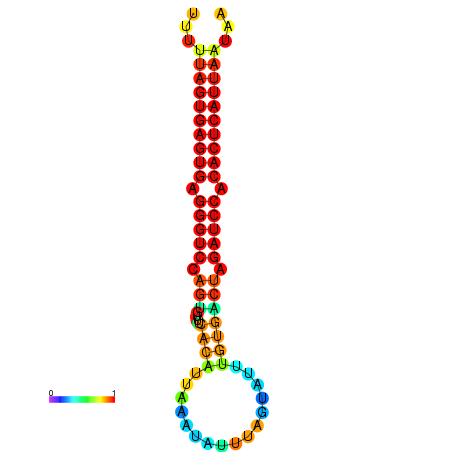| AGTGGTT-TTAAGTTCGCGATCATACTACTGTTTTTAGTGAGTGAGGGTCCAGTGTTTCACATTGATTTTCTTAGTATTTGTGACTAGATCCACACTCATTAATAACGGTAGTTCAATCAACAGAATGCACGT | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| .................................................................................TGACTAGATCCACACTCATT................................ | 20 | 1 | 2474.00 | 2474 | 1142 | 1332 |
| .................................................................................TGACTAGATCCACACTCATTA............................... | 21 | 1 | 1955.00 | 1955 | 1611 | 344 |
| .................................................................................TGACTAGATCCACACTCATTAA.............................. | 22 | 1 | 1714.00 | 1714 | 1511 | 203 |
| .................................................................................TGACTAGATCCACACTCAT................................. | 19 | 1 | 435.00 | 435 | 216 | 219 |
| ....................................AGTGAGTGAGGGTCCAGTGTTTCACA....................................................................... | 26 | 1 | 322.00 | 322 | 302 | 20 |
| .....................................GTGAGTGAGGGTCCAGTGTTTCACA....................................................................... | 25 | 1 | 89.00 | 89 | 80 | 9 |
| .....................................GTGAGTGAGGGTCCAGTGTTTCA......................................................................... | 23 | 1 | 45.00 | 45 | 24 | 21 |
| ....................................AGTGAGTGAGGGTCCAGTGTTTCAC........................................................................ | 25 | 1 | 26.00 | 26 | 26 | 0 |
| .....................................GTGAGTGAGGGTCCAGTGTTTCAC........................................................................ | 24 | 1 | 25.00 | 25 | 20 | 5 |
| ....................................AGTGAGTGAGGGTCCAGTG.............................................................................. | 19 | 1 | 22.00 | 22 | 1 | 21 |
| ....................................AGTGAGTGAGGGTCCAGTGTTTCA......................................................................... | 24 | 1 | 20.00 | 20 | 18 | 2 |
| .....................................GTGAGTGAGGGTCCAGTGTT............................................................................ | 20 | 1 | 12.00 | 12 | 1 | 11 |
| ....................................AGTGAGTGAGGGTCCAGTGT............................................................................. | 20 | 1 | 10.00 | 10 | 1 | 9 |
| .................................................................................TGACTAGATCCACACTCATTAAT............................. | 23 | 1 | 10.00 | 10 | 10 | 0 |
| ....................................AGTGAGTGAGGGTCCAGTGTTTC.......................................................................... | 23 | 1 | 9.00 | 9 | 8 | 1 |
| .....................................GTGAGTGAGGGTCCAGTGTTTC.......................................................................... | 22 | 1 | 9.00 | 9 | 3 | 6 |
| ....................................AGTGAGTGAGGGTCCAGTGTTT........................................................................... | 22 | 1 | 8.00 | 8 | 7 | 1 |
| .....................................GTGAGTGAGGGTCCAGTGTTT........................................................................... | 21 | 1 | 5.00 | 5 | 1 | 4 |
| ....................................AGTGAGTGAGGGTCCAGTGTT............................................................................ | 21 | 1 | 5.00 | 5 | 0 | 5 |
| ..............................................................TTGATTTTCTTAGTATTTG.................................................... | 19 | 1 | 3.00 | 3 | 3 | 0 |
| ....................................................................................CTAGATCCACACTCATTA............................... | 18 | 1 | 2.00 | 2 | 2 | 0 |
| ....................................AGTGAGTGAGGGTCCAGT............................................................................... | 18 | 1 | 2.00 | 2 | 1 | 1 |
| .....................................GTGAGTGAGGGTCCAGTGTTTCACAT...................................................................... | 26 | 1 | 2.00 | 2 | 2 | 0 |
| .....................................GTGAGTGAGGGTCCAGTG.............................................................................. | 18 | 1 | 2.00 | 2 | 0 | 2 |
| ..................................................................................GACTAGATCCACACTCATT................................ | 19 | 1 | 1.00 | 1 | 0 | 1 |
| .................................................................................TGACTAGATCCACACTCA.................................. | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ..................................................................................GACTAGATCCACACTCATTAA.............................. | 21 | 1 | 1.00 | 1 | 0 | 1 |