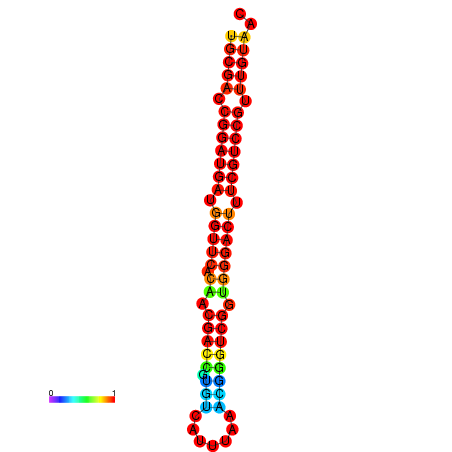
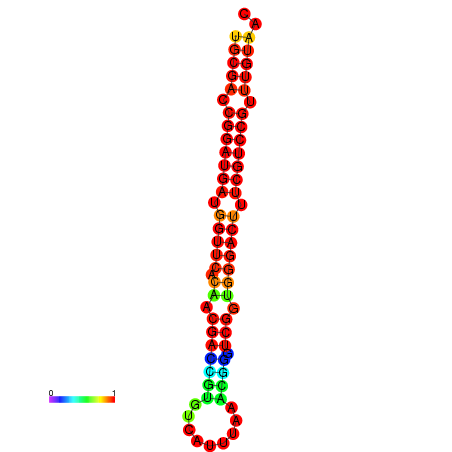
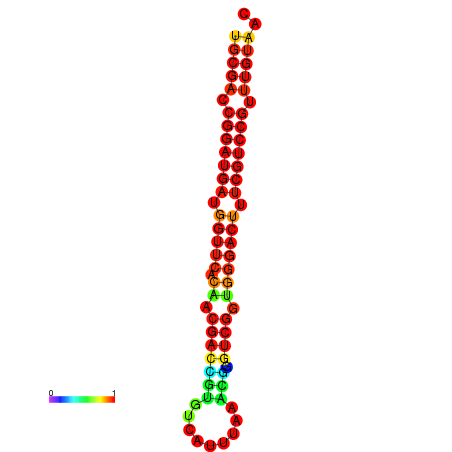
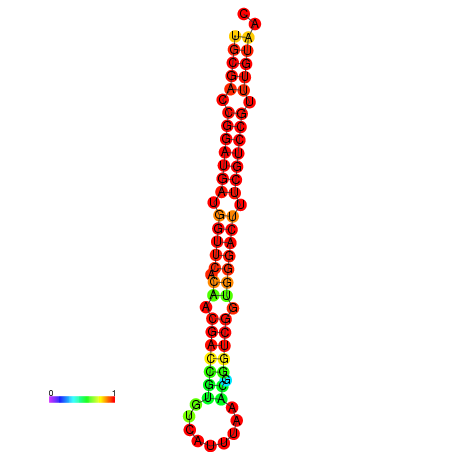
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:11544556-11544683 + | T--GATACGAG---------------------------ACCATGGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCATTTAAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------G---------TGTCCGT |
| droSim1 | chr2R:10277194-10277321 + | T--GATACGAG---------------------------ACCATGGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCCTTTAAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------G---------TGTCCGT |
| droSec1 | super_1:9037606-9037733 + | T--GATACGAG---------------------------ACCATAGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCCTTTAAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------G---------TGTCCGT |
| droYak2 | chr2R:17445962-17446088 - | T--GATACGAC---------------------------ACCATGGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCCTTTAAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------G---------TGTCCG- |
| droEre2 | scaffold_4845:14318899-14319025 - | T--GATACGAG---------------------------ACCATGGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCCTTTAAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------G---------TGTCCG- |
| droAna3 | scaffold_13266:12641317-12641435 - | TATG-----------------------------------TAACGGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCCTTCGAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAGCGATTTATCA----------------------------TCCGG |
| dp4 | chr3:19640883-19641018 - | T--G-----------------------------------CAACGGTAATGGTACGGTGCGACCGGACGATGGTTCACAACGACCGTG---TCTCTTAAACTGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAGCGATTTATCATCATCGTCGCGCCCCAG---------TGCTCGT |
| droPer1 | super_37:642280-642415 - | T--G-----------------------------------CAACGGTAATGGTACGGTGCGACCGGACGATGGTTCACAACGACCGTG---TCTCTTAAACTGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAGCGATTTATCATCATCGTCGCGCCCCAG---------TGCTCGT |
| droWil1 | scaffold_181009:1555699-1555856 - | T--GTAACGAGTTAAAAAAAAATAAGGGGAGATCCCCCATAATGGTAATGGTACGGTGCGACCGGATGACGGTTCACAACGACCGTG---TCAATTTTACTGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCATAATC----------------------TCTAGG |
| droVir3 | scaffold_12875:6427523-6427640 - | T--G---------------------------------------GTTTATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTGCA-TACTTTTAACTGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------A---ATCAG------AT |
| droMoj3 | scaffold_6496:23589388-23589515 + | T--G---------------------------------------GTTTATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTACAGTTTTTTCAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------GCAAATCAGATATCAGC |
| droGri2 | scaffold_15112:1314106-1314223 - | T--GGTC--------------------------------------TGATGGTACGGTGCGACCGGATGACGGTTCACAACGACCGTGCT-TATTTTTAACTGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAGCGATTTATCA----------------A----------ATCAGT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:11544556-11544683 + | T--GATACGAG---------------------------ACCATGGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCATTTAAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------G---------TGTCCGT |
| droSim1 | chr2R:10277194-10277321 + | T--GATACGAG---------------------------ACCATGGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCCTTTAAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------G---------TGTCCGT |
| droSec1 | super_1:9037606-9037733 + | T--GATACGAG---------------------------ACCATAGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCCTTTAAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------G---------TGTCCGT |
| droYak2 | chr2R:17445962-17446088 - | T--GATACGAC---------------------------ACCATGGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCCTTTAAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------G---------TGTCCG- |
| droEre2 | scaffold_4845:14318899-14319025 - | T--GATACGAG---------------------------ACCATGGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCCTTTAAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------G---------TGTCCG- |
| droAna3 | scaffold_13266:12641317-12641435 - | TATG-----------------------------------TAACGGTAATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTG---TCCTTCGAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAGCGATTTATCA----------------------------TCCGG |
| dp4 | chr3:19640883-19641018 - | T--G-----------------------------------CAACGGTAATGGTACGGTGCGACCGGACGATGGTTCACAACGACCGTG---TCTCTTAAACTGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAGCGATTTATCATCATCGTCGCGCCCCAG---------TGCTCGT |
| droPer1 | super_37:642280-642415 - | T--G-----------------------------------CAACGGTAATGGTACGGTGCGACCGGACGATGGTTCACAACGACCGTG---TCTCTTAAACTGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAGCGATTTATCATCATCGTCGCGCCCCAG---------TGCTCGT |
| droWil1 | scaffold_181009:1555699-1555856 - | T--GTAACGAGTTAAAAAAAAATAAGGGGAGATCCCCCATAATGGTAATGGTACGGTGCGACCGGATGACGGTTCACAACGACCGTG---TCAATTTTACTGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCATAATC----------------------TCTAGG |
| droVir3 | scaffold_12875:6427523-6427640 - | T--G---------------------------------------GTTTATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTGCA-TACTTTTAACTGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------A---ATCAG------AT |
| droMoj3 | scaffold_6496:23589388-23589515 + | T--G---------------------------------------GTTTATGGTACGGTGCGACCGGATGATGGTTCACAACGACCGTACAGTTTTTTCAACGGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAACGATTTATCA----------------GCAAATCAGATATCAGC |
| droGri2 | scaffold_15112:1314106-1314223 - | T--GGTC--------------------------------------TGATGGTACGGTGCGACCGGATGACGGTTCACAACGACCGTGCT-TATTTTTAACTGGTCGGTGGGACTTTCGTCCGTTTGTAACGCCATTTGTCAGCGATTTATCA----------------A----------ATCAGT |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-30.8, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
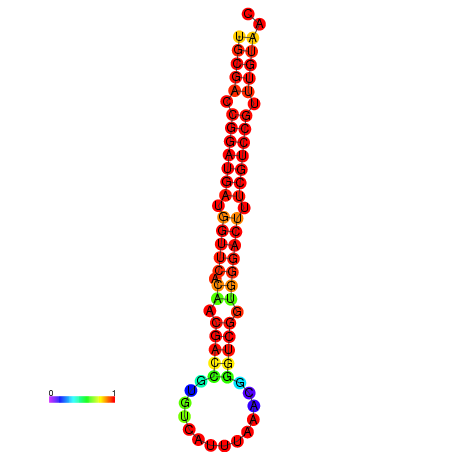
| dG=-30.8, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
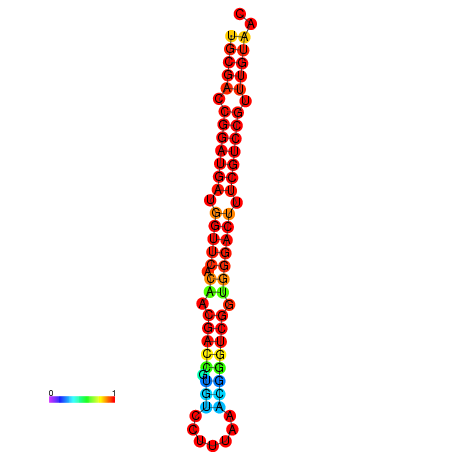 |
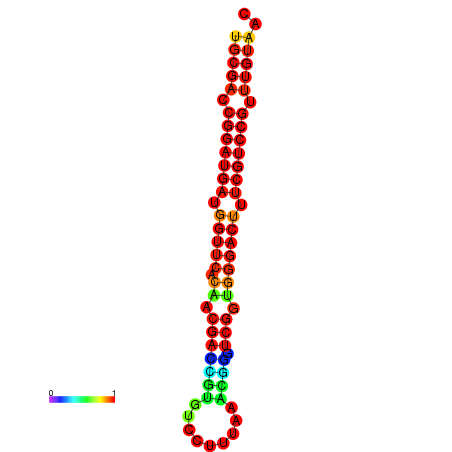 |
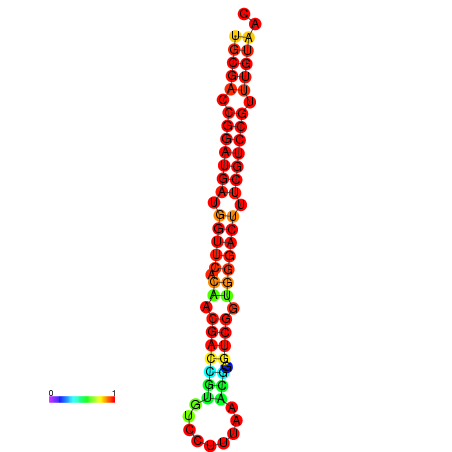 |
 |
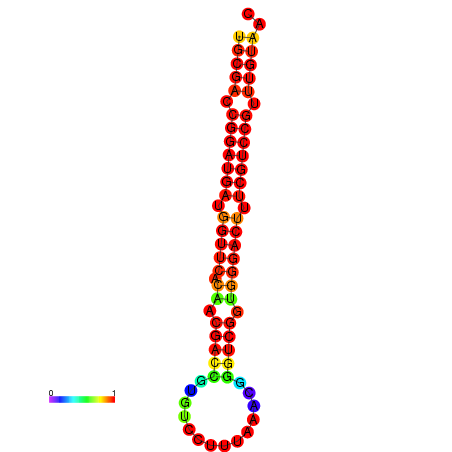 |
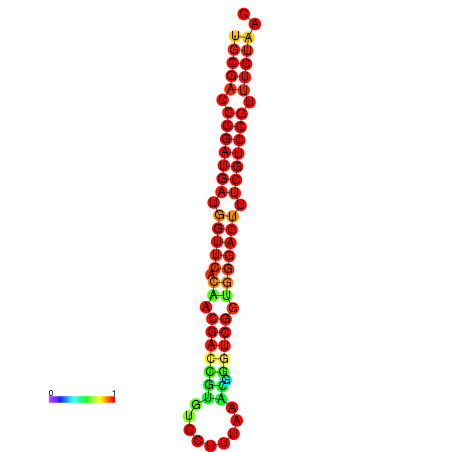
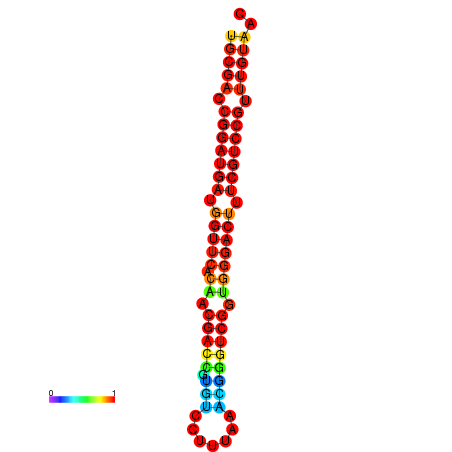
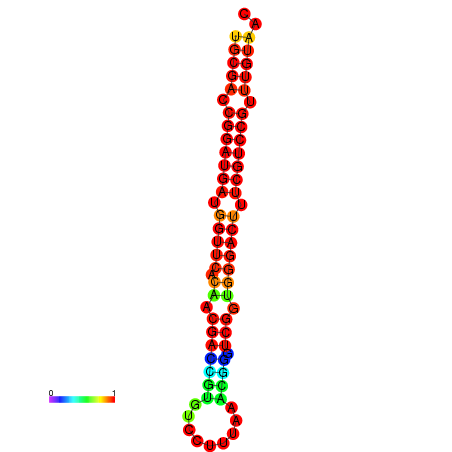
| dG=-30.8, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
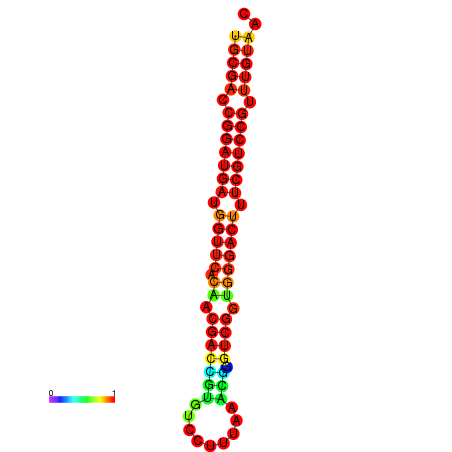 |
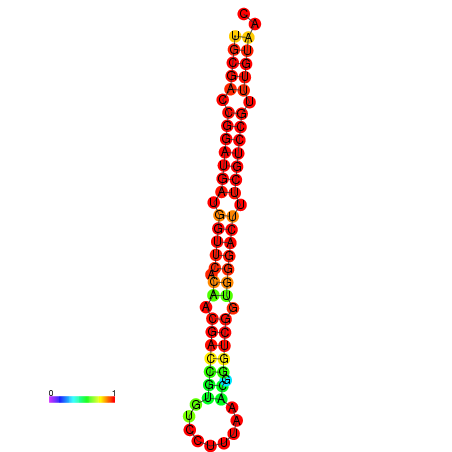 |
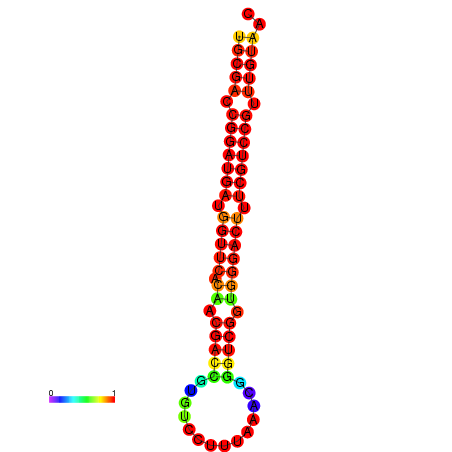 |
| dG=-30.8, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
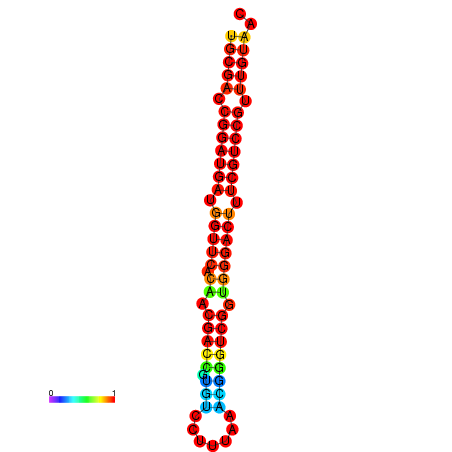 |
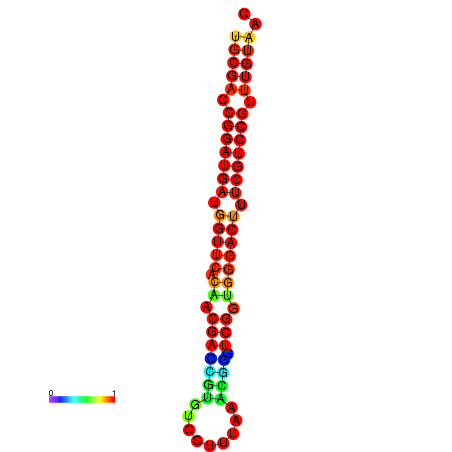 |
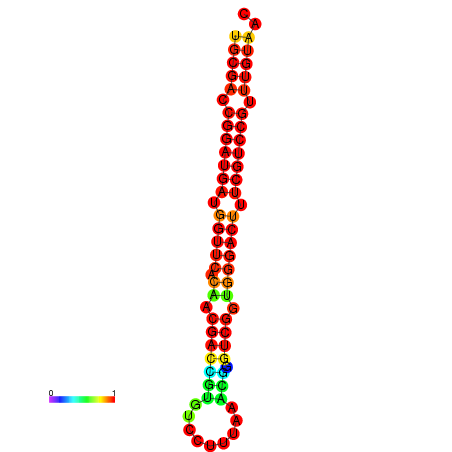 |
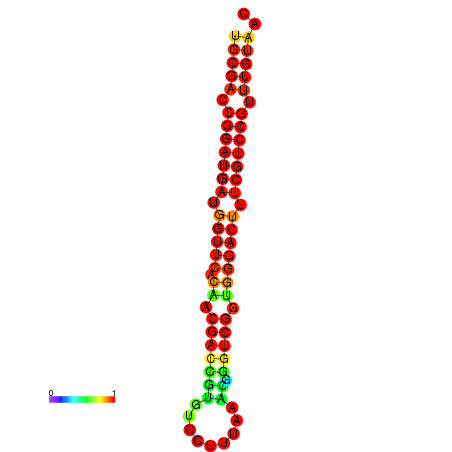 |
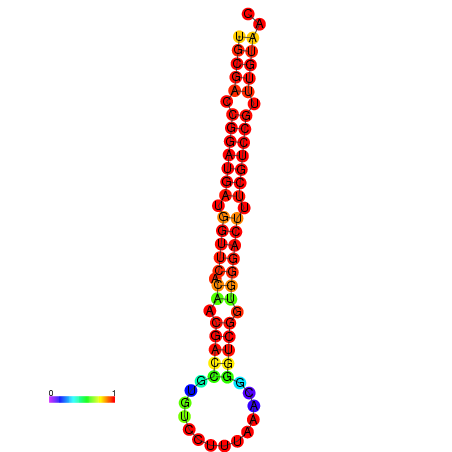 |
| dG=-30.8, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.6, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
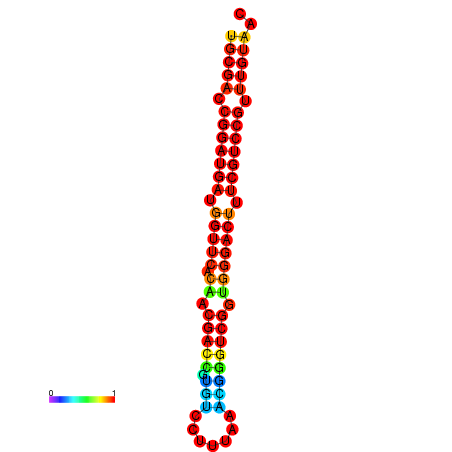 |
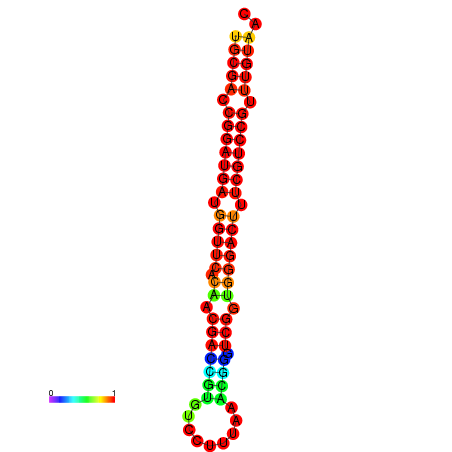 |
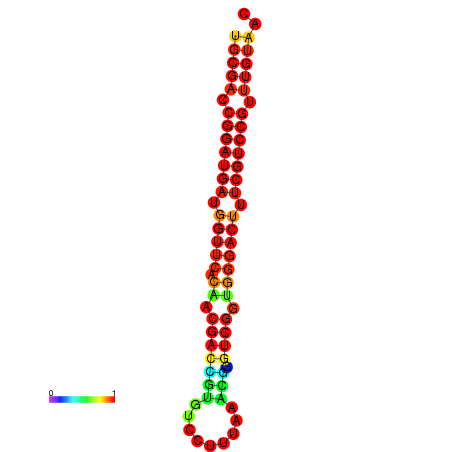 |
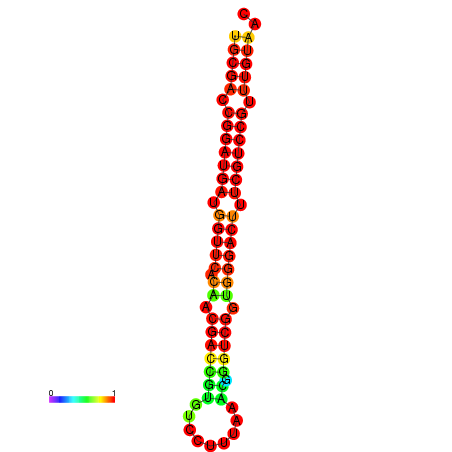 |
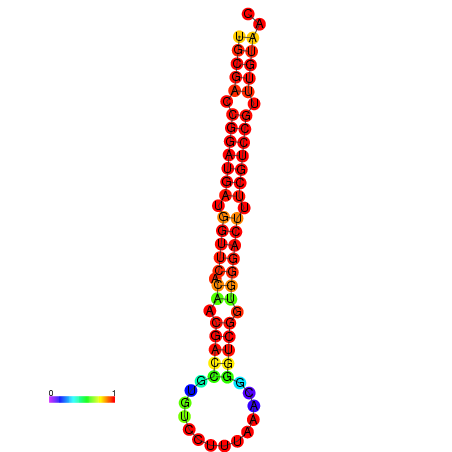 |
| dG=-31.7, p-value=0.009901 | dG=-30.8, p-value=0.009901 | dG=-30.7, p-value=0.009901 |
|---|---|---|
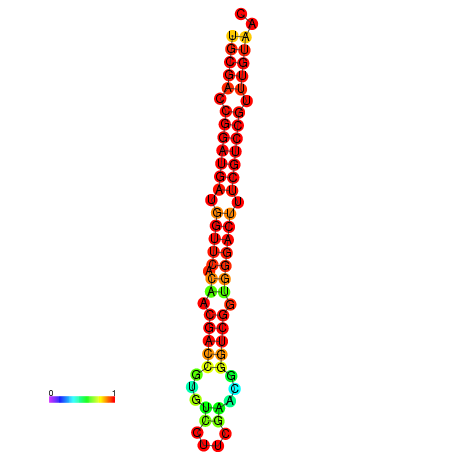 |
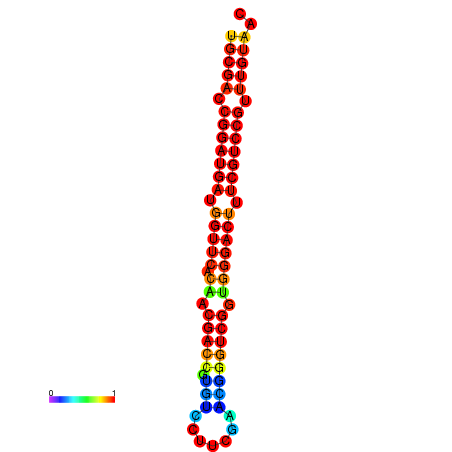 |
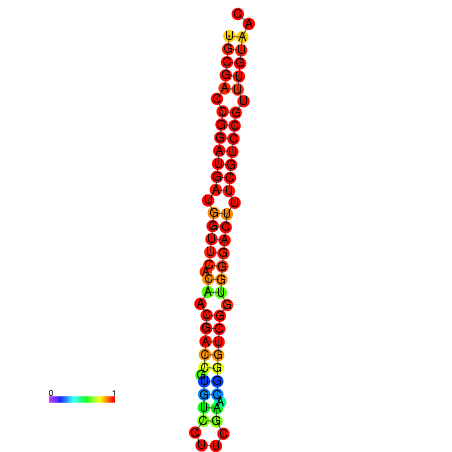 |
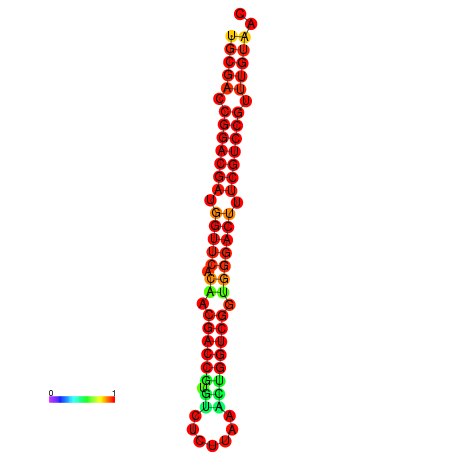
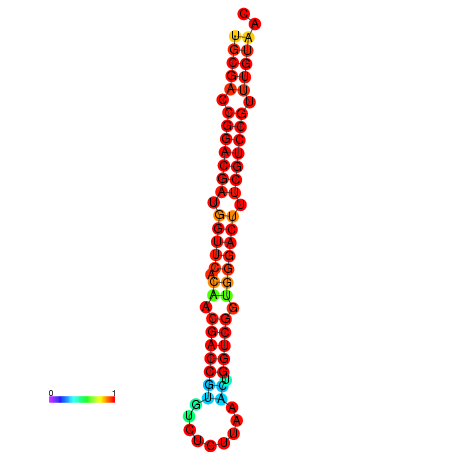
| dG=-32.6, p-value=0.009901 | dG=-32.4, p-value=0.009901 |
|---|---|
 |
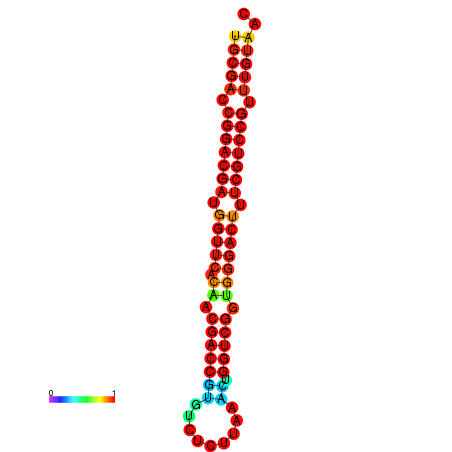 |
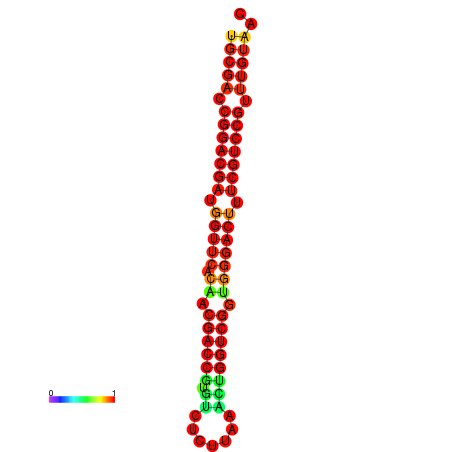
| dG=-32.6, p-value=0.009901 | dG=-32.4, p-value=0.009901 |
|---|---|
 |
 |
| dG=-31.2, p-value=0.009901 | dG=-30.3, p-value=0.009901 |
|---|---|
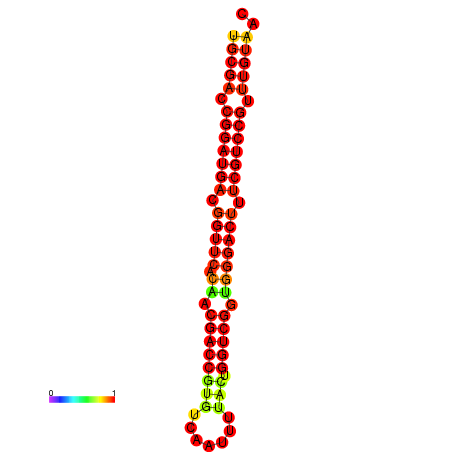 |
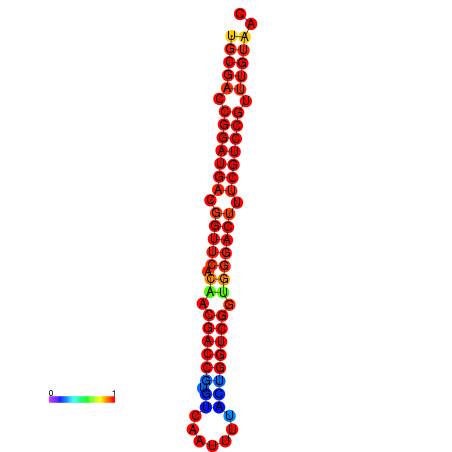 |
| dG=-30.4, p-value=0.009901 | dG=-29.5, p-value=0.009901 |
|---|---|
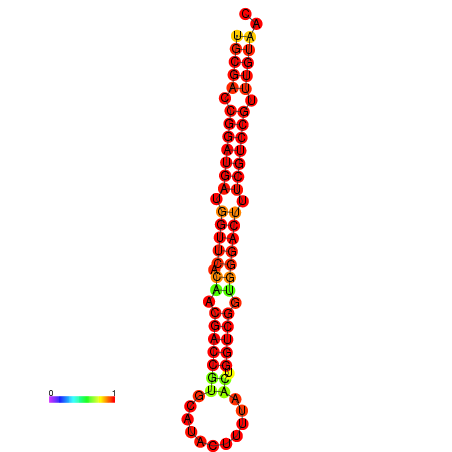 |
 |
| dG=-30.0, p-value=0.009901 | dG=-29.3, p-value=0.009901 | dG=-29.3, p-value=0.009901 | dG=-29.3, p-value=0.009901 |
|---|---|---|---|
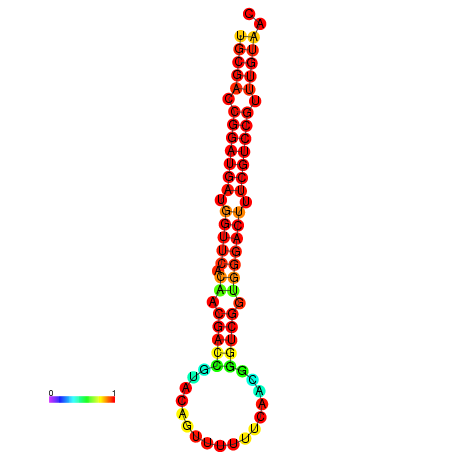 |
 |
 |
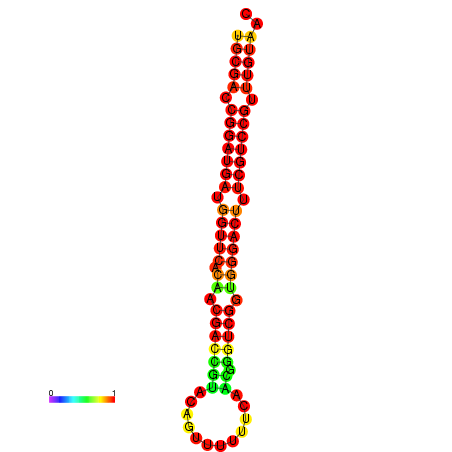 |
| dG=-30.1, p-value=0.009901 | dG=-29.9, p-value=0.009901 | dG=-29.2, p-value=0.009901 |
|---|---|---|
 |
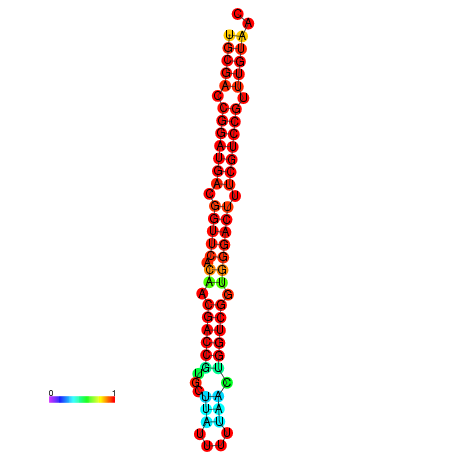 |
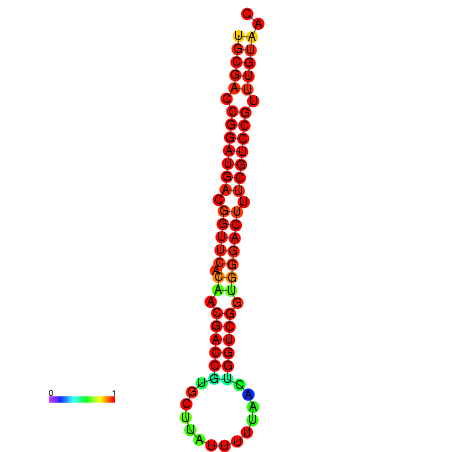 |
Generated: 03/07/2013 at 05:02 PM