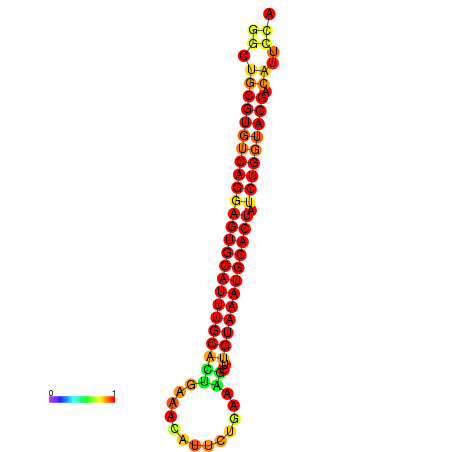
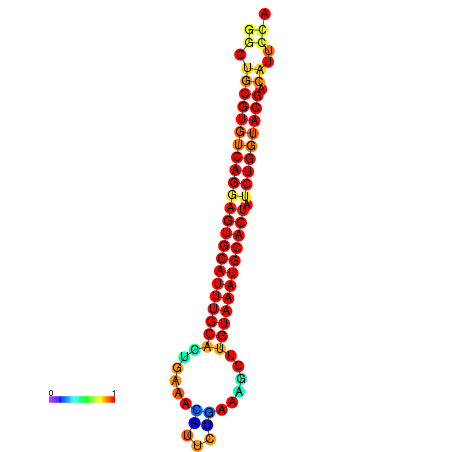
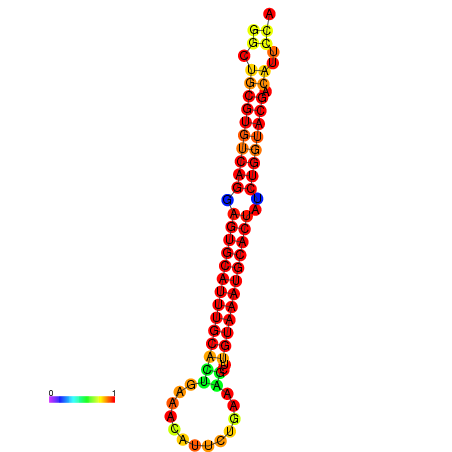
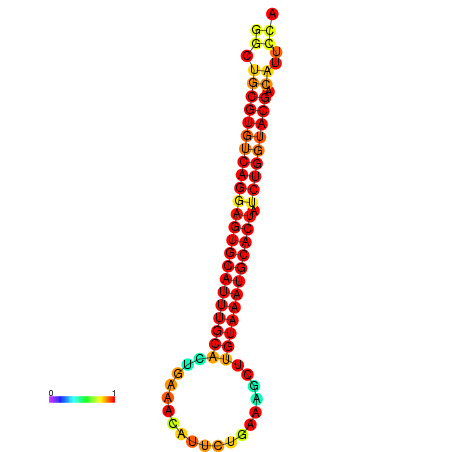| GATTAACTGAGCATCCTTTGAAGGTTTTGGGTTGCGTGTCAGGAGTGCATTTGCACTGAAACATTCTGAAGCTTGTAAATGCACTATCTGGTACGACATTCCAGAACGTACAATCTTCCCGAATGCAAGACAAATATGCGA | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ...........................................................................TAAATGCACTATCTGGTACGACA........................................... | 23 | 1 | 79776.00 | 79776 | 3469 | 76307 |
| ...........................................................................TAAATGCACTATCTGGTACGA............................................. | 21 | 1 | 6883.00 | 6883 | 749 | 6134 |
| ...........................................................................TAAATGCACTATCTGGTACGAC............................................ | 22 | 1 | 5851.00 | 5851 | 472 | 5379 |
| ...........................................................................TAAATGCACTATCTGGTACG.............................................. | 20 | 1 | 2200.00 | 2200 | 115 | 2085 |
| ..................................CGTGTCAGGAGTGCATTTGCA...................................................................................... | 21 | 1 | 872.00 | 872 | 694 | 178 |
| ..................................CGTGTCAGGAGTGCATTTGC....................................................................................... | 20 | 1 | 117.00 | 117 | 25 | 92 |
| ............................................................................AAATGCACTATCTGGTACGACA........................................... | 22 | 1 | 115.00 | 115 | 5 | 110 |
| ...........................................................................TAAATGCACTATCTGGTACGACAT.......................................... | 24 | 1 | 58.00 | 58 | 1 | 57 |
| .............................................................................AATGCACTATCTGGTACGACA........................................... | 21 | 1 | 43.00 | 43 | 4 | 39 |
| ...................................GTGTCAGGAGTGCATTTGCA...................................................................................... | 20 | 1 | 26.00 | 26 | 19 | 7 |
| ...........................................................................TAAATGCACTATCTGGTAC............................................... | 19 | 1 | 26.00 | 26 | 3 | 23 |
| ..............................................................................ATGCACTATCTGGTACGACA........................................... | 20 | 1 | 24.00 | 24 | 1 | 23 |
| ...............................................................................TGCACTATCTGGTACGACA........................................... | 19 | 1 | 22.00 | 22 | 7 | 15 |
| ................................................................................GCACTATCTGGTACGACA........................................... | 18 | 1 | 18.00 | 18 | 7 | 11 |
| .......................................................CTGAAACATTCTGAAGCTTG.................................................................. | 20 | 1 | 16.00 | 16 | 5 | 11 |
| ............................................................................AAATGCACTATCTGGTACGAC............................................ | 21 | 1 | 6.00 | 6 | 1 | 5 |
| ..................................CGTGTCAGGAGTGCATTTG........................................................................................ | 19 | 1 | 6.00 | 6 | 1 | 5 |
| ............................................................................AAATGCACTATCTGGTACGA............................................. | 20 | 1 | 5.00 | 5 | 0 | 5 |
| ..................................CGTGTCAGGAGTGCATTTGCACTGAA................................................................................. | 26 | 1 | 5.00 | 5 | 1 | 4 |
| .............................................................................AATGCACTATCTGGTACGAC............................................ | 20 | 1 | 5.00 | 5 | 0 | 5 |