| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:10312870-10312998 + | GACCA-------TC-----GAAC-GA--AAAAAACCGAAGTCTTTTTACCATCAGCGAGGTATAGAGTTCCTACGTTCCTATATT-C---AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGACTGTCGACCAGCATA--------TAGTTAAAA |
| droSim1 | chr3L:9702897-9703022 + | GACCA-------TC-----GAAC-G---AAAAAACCGAAGTCTTTTTACCATCAGCGAGGTATAGAGTTCCTACGTTCCT--ATT-C---AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGACTGTCGACCATCATA--------TAGTTGAAA |
| droSec1 | super_0:2526747-2526871 + | GACCA--------C-----GAAC-G---AAAAAACCGAAGTCTTTTTACCATCAGCGAGGTATAGAGTTCCTACGTTCCT--ATT-C---AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGACTGTCGACCATCATA--------TAGTTGAAA |
| droYak2 | chr3L:10302128-10302256 + | GACAA-------CA-----GAAA-GAACAAAAAACCGAAGTCTTTTTACCATCAGCGAGGTATAGAGTTCCTACGTTCCT--ATT-C---AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGACTGTCGACCATCATA--------AAGTTAAAA |
| droEre2 | scaffold_4784:10306167-10306292 + | GACAA-------CC-----GAAT-A---AAAAAACCGAAGTCTTTTTACCATCAGCGAGGTATAGAGTTCCTACGTTCCT--ATT-C---AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGACTGTCGACCATCATA--------AAGTTGAAA |
| droAna3 | scaffold_13337:15233368-15233487 + | ATTT------------------CTG---AAGAAACCGAAGCCTATTTGCCATCAGCGAGGTATAGAGTTCCTACGTGCCA--CTTAAACTAGTCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCGACCATCATG--------A-----AAG |
| dp4 | chrXR_group6:8706029-8706135 + | ATCTA---------------------------AAGCGAAGCCTCTTTACCATCAGCGAGGTATAGAGTTCCTACGTGCCG-TATC-CAA-AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCCATCAAC-------------------- |
| droPer1 | super_27:1146104-1146210 + | ATCTA---------------------------AAGCGAAGCCTCTTTACCATCAGCGAGGTATAGAGTTCCTACGTGCCG-TATC-CAA-AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCCATCAAC-------------------- |
| droWil1 | scaffold_181024:290398-290516 - | AACAA--------------ATTT-G---AAATAAACGAAGCCTATTTACCATAAGCGAGGTATAGAGTTCCTACGTGCAA--ATTGCAATAATCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCGAACAACAA-----------------A |
| droVir3 | scaffold_13049:16093198-16093326 + | TATCT--------T---AAGATTTA---AAGATACCGAAGTCTATATACCATCAGCGAGGTATAGAGTTCCTACGTGCAA--ATTGCAATAATCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCGACCAGCAA-----------GAAAAGA |
| droMoj3 | scaffold_6680:20678203-20678343 - | GACCAAGTGAAACAGAG--ATTGTA---AAGATAACGAAGTCTATATACCATCAGCGAGGTATAGAGTTCCTACGTGCAA--ATTGCAATAGTCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCGACCAACAAA--------ACGAATAAA |
| droGri2 | scaffold_15110:12572308-12572448 - | AAATC-------TC---AAAACCGA---AAGATAACGAAGTCTATTTACCATTAGCGAGGTATAGAGTTCCTACGTGCAA--ATTGCAATAATCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCGACCAGCATTGATCAAAACAATTAAGT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:10312870-10312998 + | GACCA-------TC-----GAAC-GA--AAAAAACCGAAGTCTTTTTACCATCAGCGAGGTATAGAGTTCCTACGTTCCTATATT-C---AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGACTGTCGACCAGCATA--------TAGTTAAAA |
| droSim1 | chr3L:9702897-9703022 + | GACCA-------TC-----GAAC-G---AAAAAACCGAAGTCTTTTTACCATCAGCGAGGTATAGAGTTCCTACGTTCCT--ATT-C---AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGACTGTCGACCATCATA--------TAGTTGAAA |
| droSec1 | super_0:2526747-2526871 + | GACCA--------C-----GAAC-G---AAAAAACCGAAGTCTTTTTACCATCAGCGAGGTATAGAGTTCCTACGTTCCT--ATT-C---AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGACTGTCGACCATCATA--------TAGTTGAAA |
| droYak2 | chr3L:10302128-10302256 + | GACAA-------CA-----GAAA-GAACAAAAAACCGAAGTCTTTTTACCATCAGCGAGGTATAGAGTTCCTACGTTCCT--ATT-C---AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGACTGTCGACCATCATA--------AAGTTAAAA |
| droEre2 | scaffold_4784:10306167-10306292 + | GACAA-------CC-----GAAT-A---AAAAAACCGAAGTCTTTTTACCATCAGCGAGGTATAGAGTTCCTACGTTCCT--ATT-C---AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGACTGTCGACCATCATA--------AAGTTGAAA |
| droAna3 | scaffold_13337:15233368-15233487 + | ATTT------------------CTG---AAGAAACCGAAGCCTATTTGCCATCAGCGAGGTATAGAGTTCCTACGTGCCA--CTTAAACTAGTCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCGACCATCATG--------A-----AAG |
| dp4 | chrXR_group6:8706029-8706135 + | ATCTA---------------------------AAGCGAAGCCTCTTTACCATCAGCGAGGTATAGAGTTCCTACGTGCCG-TATC-CAA-AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCCATCAAC-------------------- |
| droPer1 | super_27:1146104-1146210 + | ATCTA---------------------------AAGCGAAGCCTCTTTACCATCAGCGAGGTATAGAGTTCCTACGTGCCG-TATC-CAA-AGTCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCCATCAAC-------------------- |
| droWil1 | scaffold_181024:290398-290516 - | AACAA--------------ATTT-G---AAATAAACGAAGCCTATTTACCATAAGCGAGGTATAGAGTTCCTACGTGCAA--ATTGCAATAATCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCGAACAACAA-----------------A |
| droVir3 | scaffold_13049:16093198-16093326 + | TATCT--------T---AAGATTTA---AAGATACCGAAGTCTATATACCATCAGCGAGGTATAGAGTTCCTACGTGCAA--ATTGCAATAATCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCGACCAGCAA-----------GAAAAGA |
| droMoj3 | scaffold_6680:20678203-20678343 - | GACCAAGTGAAACAGAG--ATTGTA---AAGATAACGAAGTCTATATACCATCAGCGAGGTATAGAGTTCCTACGTGCAA--ATTGCAATAGTCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCGACCAACAAA--------ACGAATAAA |
| droGri2 | scaffold_15110:12572308-12572448 - | AAATC-------TC---AAAACCGA---AAGATAACGAAGTCTATTTACCATTAGCGAGGTATAGAGTTCCTACGTGCAA--ATTGCAATAATCGTAGGAACTTAATACCGTGCTCTTGGAGGGCTGTCGACCAGCATTGATCAAAACAATTAAGT |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
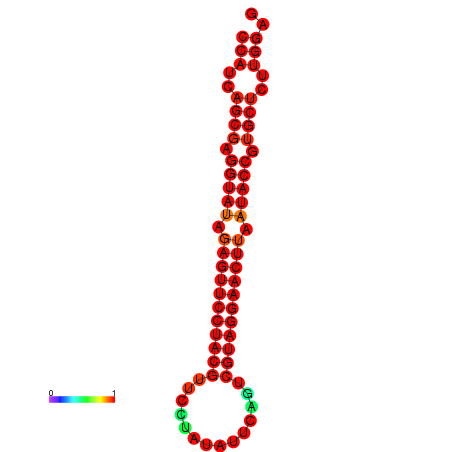
| dG=-33.4, p-value=0.009901 | dG=-33.4, p-value=0.009901 |
|---|---|
 |
 |
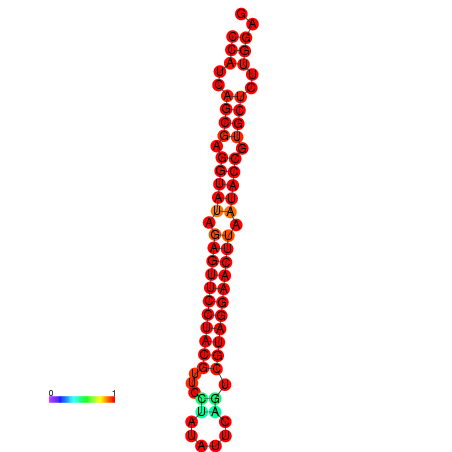
| dG=-33.6, p-value=0.009901 | dG=-33.2, p-value=0.009901 |
|---|---|
 |
 |
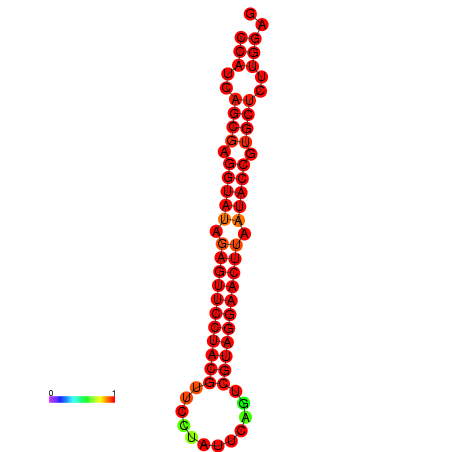
| dG=-33.6, p-value=0.009901 | dG=-33.2, p-value=0.009901 |
|---|---|
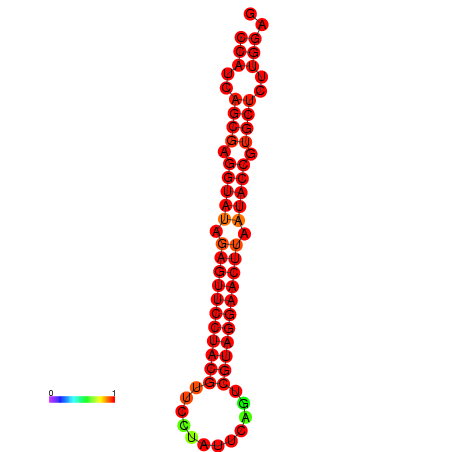 |
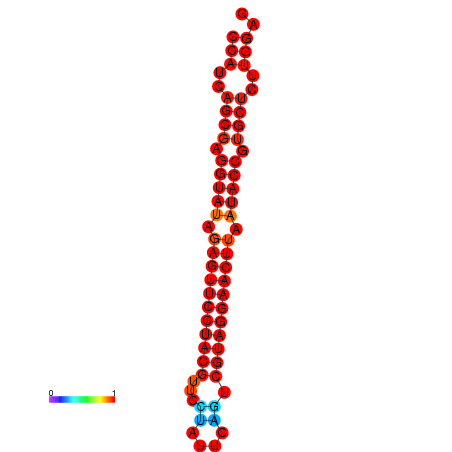 |
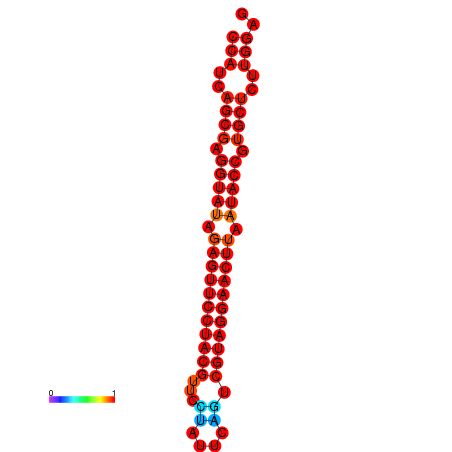
| dG=-33.6, p-value=0.009901 | dG=-33.2, p-value=0.009901 |
|---|---|
 |
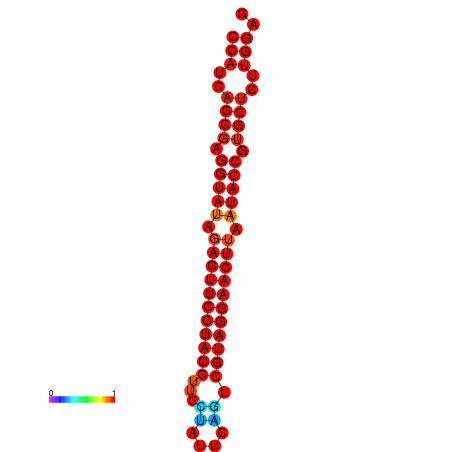 |
| dG=-33.6, p-value=0.009901 | dG=-33.2, p-value=0.009901 |
|---|---|
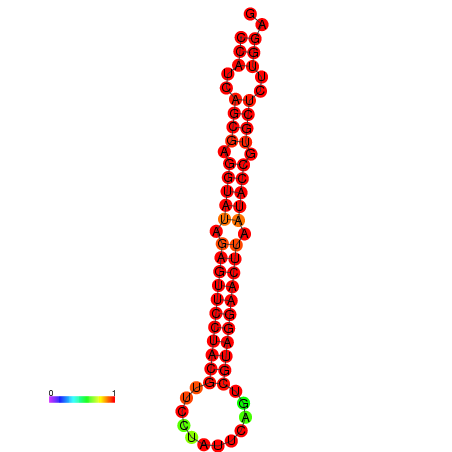 |
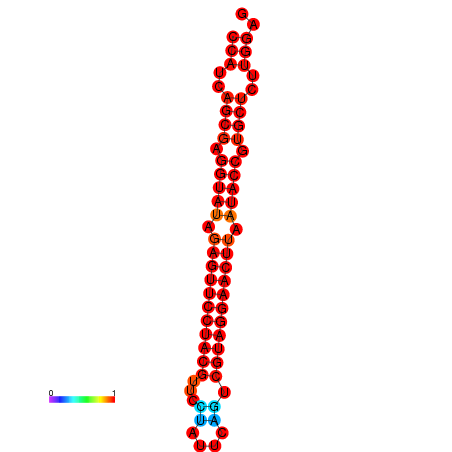 |
| dG=-34.4, p-value=0.009901 |
|---|
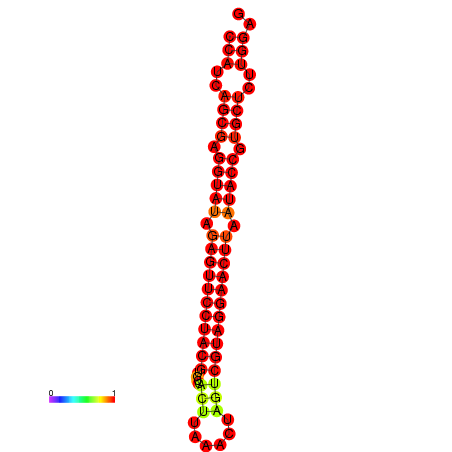 |
| dG=-33.4, p-value=0.009901 | dG=-33.4, p-value=0.009901 |
|---|---|
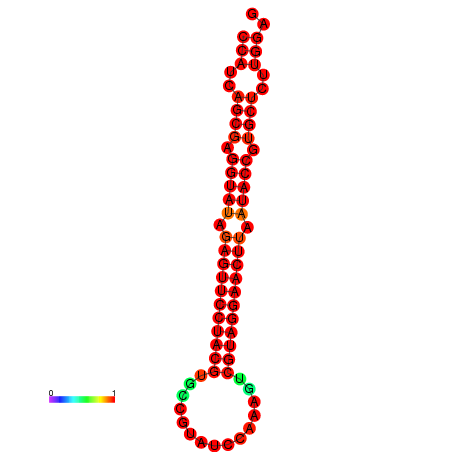 |
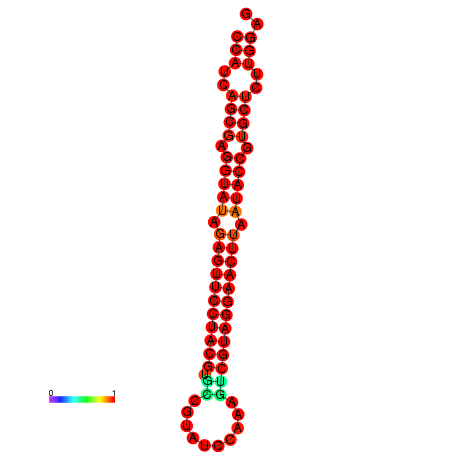 |
| dG=-33.4, p-value=0.009901 | dG=-33.4, p-value=0.009901 |
|---|---|
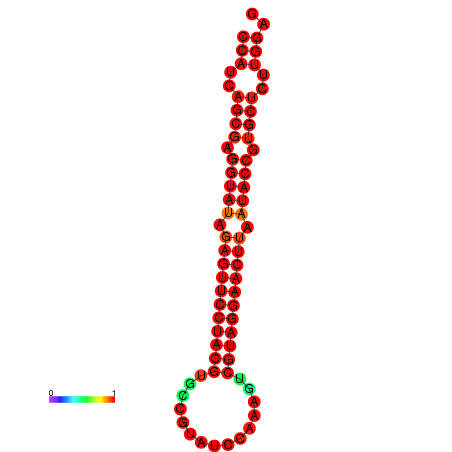 |
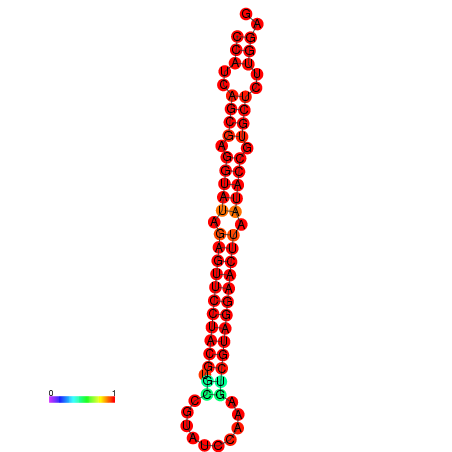 |
| dG=-36.4, p-value=0.009901 | dG=-36.0, p-value=0.009901 |
|---|---|
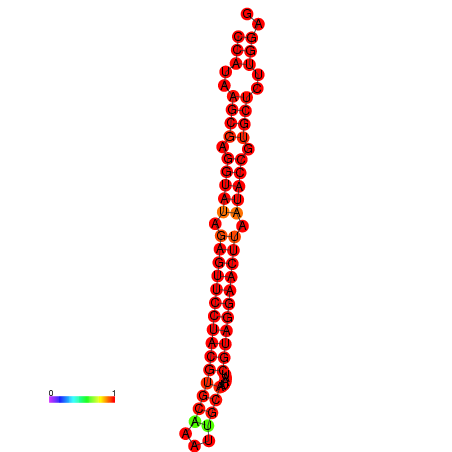 |
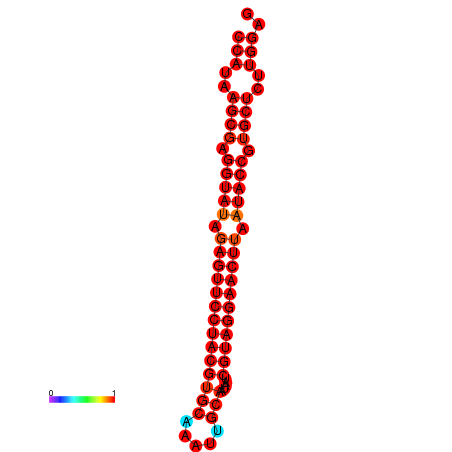 |
| dG=-35.9, p-value=0.009901 | dG=-35.5, p-value=0.009901 |
|---|---|
 |
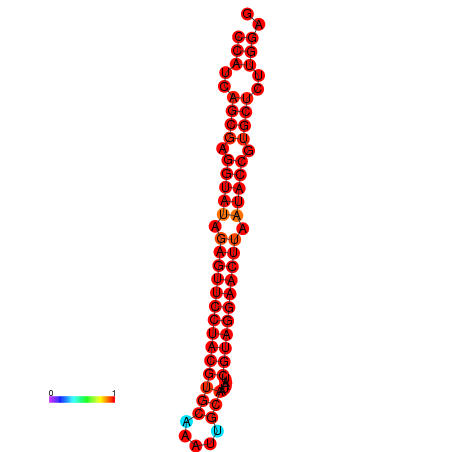 |
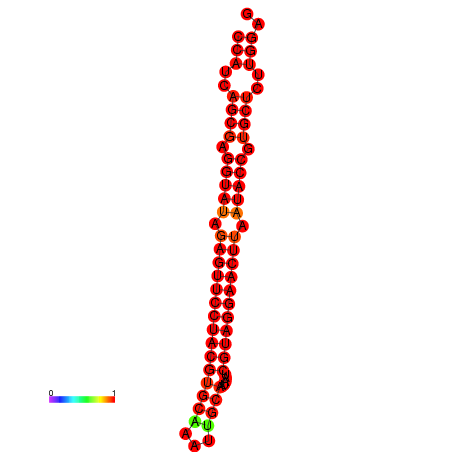
| dG=-35.9, p-value=0.009901 | dG=-35.5, p-value=0.009901 |
|---|---|
 |
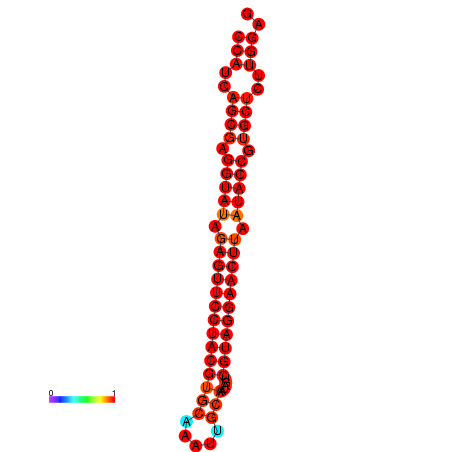 |
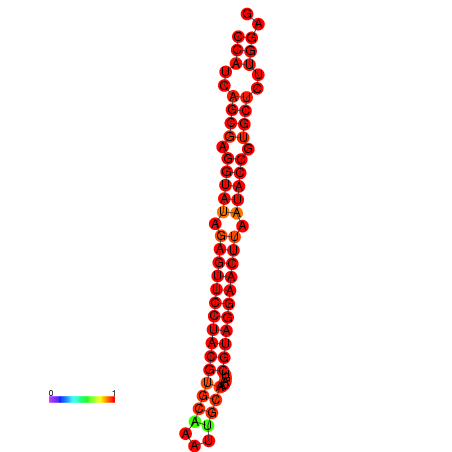
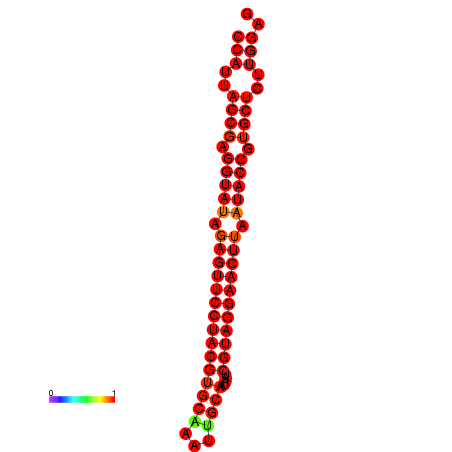
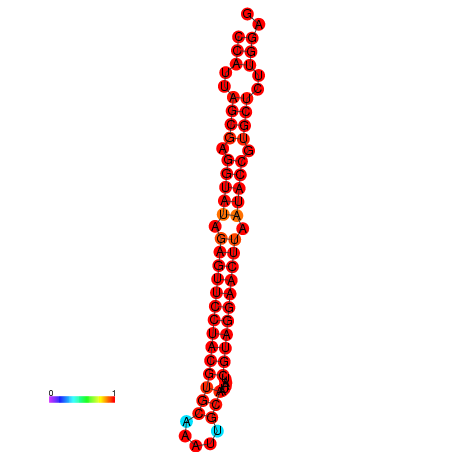
| dG=-36.2, p-value=0.009901 | dG=-35.8, p-value=0.009901 |
|---|---|
 |
 |
Generated: 03/07/2013 at 05:00 PM