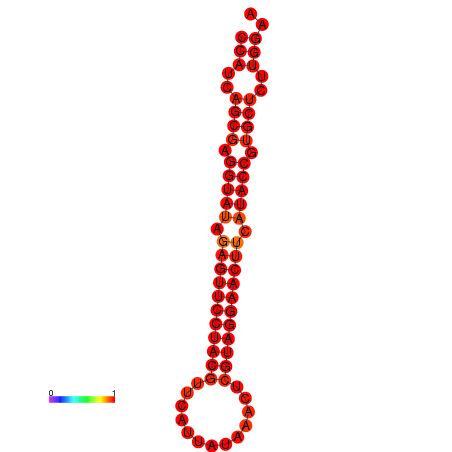
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:10358313-10358440 + | CC-AA----------GA---------TATTTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCA--TTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-CAAGTGAATGCAAAA-------------------------------------ATCC |
| droSim1 | chr3L:9747980-9748107 + | CC-AA----------GA---------TATTTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCA--TTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-CAAGTGAATGCAAAA-------------------------------------ATCC |
| droSec1 | super_0:2571151-2571278 + | CC-AA----------GA---------TATTTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCA--TTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-CAAGTGAATGCAAAA-------------------------------------ATCC |
| droYak2 | chr3L:10348871-10348998 + | GC-AA----------GA---------TATTTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCA--TTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-CAAGTGAATGCAAAA-------------------------------------ATCC |
| droEre2 | scaffold_4784:10353122-10353248 + | GC-AA----------GA---------TAT-TTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCA--TTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-CAAGTGAATGCAAAA-------------------------------------ATCC |
| droAna3 | scaffold_13337:15274238-15274391 + | CC-AA----------GA---------TATTTCTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTAACATTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCTAAAATCAAC-GA-----AA------ACATAAAGCGGAGTTGGTTTCCCCTTCAACCA-AAAAAT-C |
| dp4 | chrXR_group6:8756376-8756505 + | CG-AACGGAGGAGAAGATGTGTGTATT--TTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCATATTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAGACAAC-GA------------------------------------------------------ |
| droPer1 | super_27:1191188-1191317 + | CG-AACGGAGGAGAAGATGTGTGTATT--TTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCATATTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAGACAAC-GA------------------------------------------------------ |
| droWil1 | scaffold_181024:222598-222734 - | CTGAA----------AA---------TA--TGCTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCATTTAATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-GTTACGAAA------ACAG----------------------CAACAAGAAA-AAAC |
| droVir3 | scaffold_13049:16143973-16144087 + | TG-AA----------AA---------TAT--GCTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCAATTTATATACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-GAAGTG-------------------------------------------------- |
| droMoj3 | scaffold_6680:20621809-20621940 - | CC-AA----------AG---------TAT--GCTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCAATTTAAATACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAACACAACTCAAACAAAAACGAAAACG----------------------------------AGCG |
| droGri2 | scaffold_15110:12521926-12522042 - | CA-AA----------AA---------TAT--GCTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTAAATTTAAATACTCATAGGAACTTCATACCGTGCTCTTGGAAGACCAAAACACAAC-AAAG------------------------------------------------AAAC |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:10358313-10358440 + | CC-AA----------GA---------TATTTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCA--TTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-CAAGTGAATGCAAAA-------------------------------------ATCC |
| droSim1 | chr3L:9747980-9748107 + | CC-AA----------GA---------TATTTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCA--TTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-CAAGTGAATGCAAAA-------------------------------------ATCC |
| droSec1 | super_0:2571151-2571278 + | CC-AA----------GA---------TATTTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCA--TTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-CAAGTGAATGCAAAA-------------------------------------ATCC |
| droYak2 | chr3L:10348871-10348998 + | GC-AA----------GA---------TATTTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCA--TTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-CAAGTGAATGCAAAA-------------------------------------ATCC |
| droEre2 | scaffold_4784:10353122-10353248 + | GC-AA----------GA---------TAT-TTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCA--TTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-CAAGTGAATGCAAAA-------------------------------------ATCC |
| droAna3 | scaffold_13337:15274238-15274391 + | CC-AA----------GA---------TATTTCTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTAACATTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCTAAAATCAAC-GA-----AA------ACATAAAGCGGAGTTGGTTTCCCCTTCAACCA-AAAAAT-C |
| dp4 | chrXR_group6:8756376-8756505 + | CG-AACGGAGGAGAAGATGTGTGTATT--TTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCATATTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAGACAAC-GA------------------------------------------------------ |
| droPer1 | super_27:1191188-1191317 + | CG-AACGGAGGAGAAGATGTGTGTATT--TTTTTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCATATTATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAGACAAC-GA------------------------------------------------------ |
| droWil1 | scaffold_181024:222598-222734 - | CTGAA----------AA---------TA--TGCTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCATTTAATAAACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-GTTACGAAA------ACAG----------------------CAACAAGAAA-AAAC |
| droVir3 | scaffold_13049:16143973-16144087 + | TG-AA----------AA---------TAT--GCTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCAATTTATATACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAAAACAAC-GAAGTG-------------------------------------------------- |
| droMoj3 | scaffold_6680:20621809-20621940 - | CC-AA----------AG---------TAT--GCTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTCAATTTAAATACTCGTAGGAACTTCATACCGTGCTCTTGGAAGACCAAAACACAACTCAAACAAAAACGAAAACG----------------------------------AGCG |
| droGri2 | scaffold_15110:12521926-12522042 - | CA-AA----------AA---------TAT--GCTACCTGGTTTTTGCCATCAGCGAGGTATAGAGTTCCTACGTTAAATTTAAATACTCATAGGAACTTCATACCGTGCTCTTGGAAGACCAAAACACAAC-AAAG------------------------------------------------AAAC |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-33.5, p-value=0.009901 |
|---|
 |
| dG=-33.5, p-value=0.009901 |
|---|
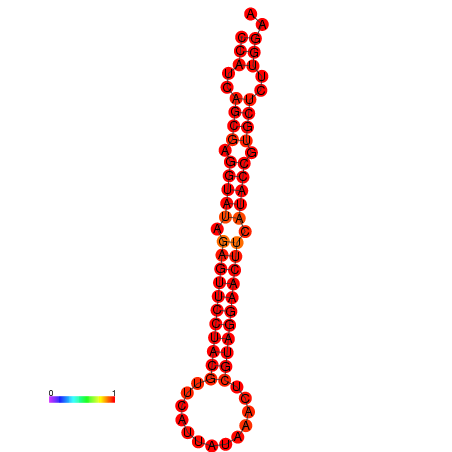 |
| dG=-33.5, p-value=0.009901 |
|---|
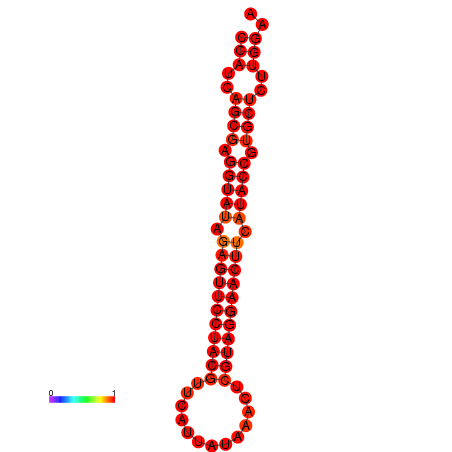 |
| dG=-33.5, p-value=0.009901 |
|---|
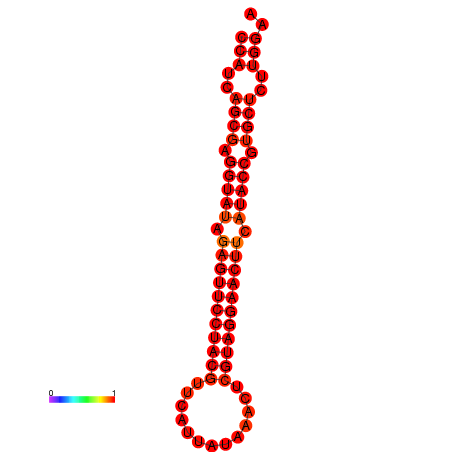 |
| dG=-33.5, p-value=0.009901 |
|---|
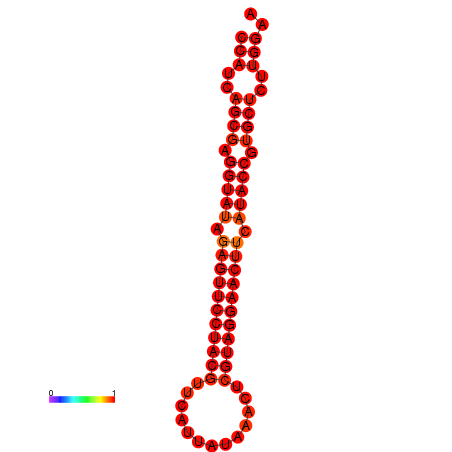 |
| dG=-33.4, p-value=0.009901 |
|---|
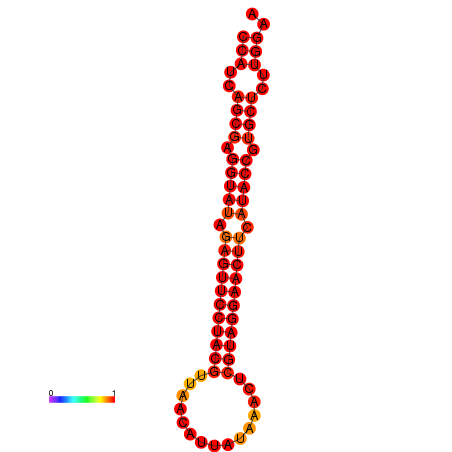 |
| dG=-33.4, p-value=0.009901 |
|---|
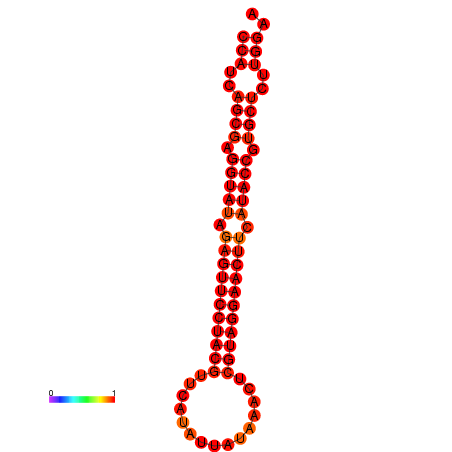 |
| dG=-33.4, p-value=0.009901 |
|---|
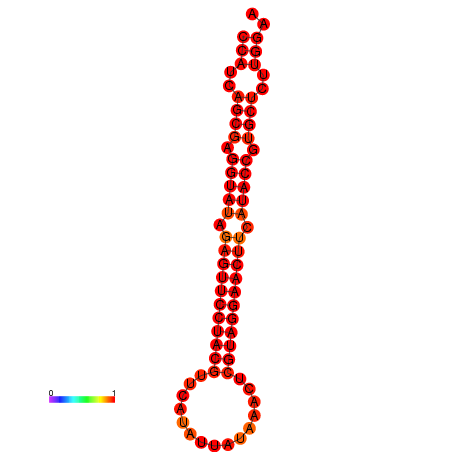 |
| dG=-33.4, p-value=0.009901 |
|---|
 |
| dG=-33.4, p-value=0.009901 |
|---|
 |
| dG=-33.4, p-value=0.009901 |
|---|
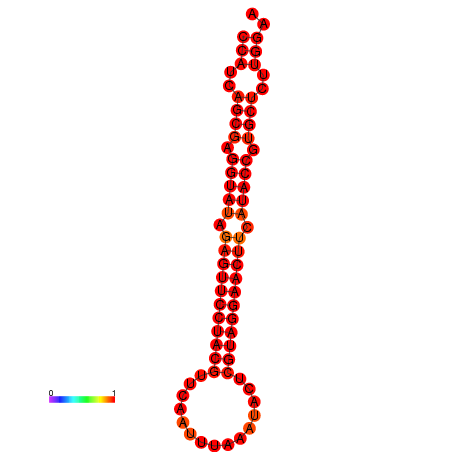 |
| dG=-27.6, p-value=0.009901 | dG=-27.0, p-value=0.009901 |
|---|---|
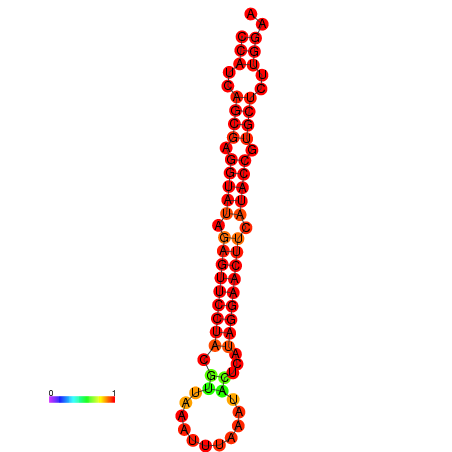 |
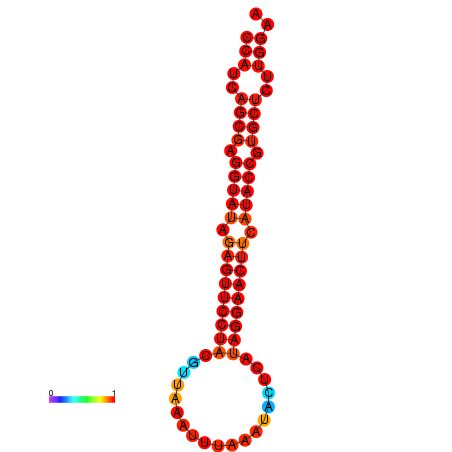 |
Generated: 03/07/2013 at 05:00 PM