| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:7425780-7425907 + | TGATGTTCCCCCGACTGTA--AAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----TTATAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGG-CAGACATATAT----CTCCATCTT-------------------CACC--AT |
| droSim1 | chr2L:7217350-7217477 + | TGATGTTCCCCCGACTGAA--AAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----TTATAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGG-CAGACATATAT----CTCCATCTC-------------------CGCC--AT |
| droSec1 | super_3:2941240-2941367 + | TGATGTTCCCCCGACTGAA--AAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTTGTTTT-----TTATAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGG-CAGACATATAT----CTCCATCTC-------------------CGTC--AT |
| droYak2 | chr2L:5473766-5473897 - | TGATGTTCCCCCGACTGAACAGAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----TTATAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGC-CAGACATATAT----CTCCAACTC-------------------CACGATAT |
| droEre2 | scaffold_4929:16344506-16344633 + | TGATGTTCCCCCGACTGAA--AAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----TTATAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGC-CAGACATATAT----CTCCATCTC-------------------CACC--AT |
| droAna3 | scaffold_12916:15333987-15334092 + | -------------ACTCTC--CAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----TGCA-----ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGG-CAGACATATAT----C---------------------------CGCC--AT |
| dp4 | chr4_group1:4175726-4175840 + | CCACCTTC-------TTGA--AAGTCTCCTACCATGCGCGCTAATCAGAGACCGGGGCTGGTTTT-----TTCACT---GCAGTCAGGTACCTGAAGTAGCGCGCGTGGTGC-TAGACACGAA--CATC--TGT----------------------------GT |
| droPer1 | super_5:1086449-1086563 - | CCACCTTC-------TTGA--AAGTCTCCTACCATGCGCGCTAATCAGAGACCGGGGCTGGTTTT-----TTCACT---GCAGTCAGGTACCTGAAGTAGCGCGCGTGGTGC-TAGACACGAA--CATC--TGT----------------------------GT |
| droWil1 | scaffold_180772:5257564-5257692 - | -----------------------GTCTCTTACCTTGCGCGCTAATCAGTGACCAGGGCTGAGTAATTTTATTCAAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTTT-TAGACCAAGATTCATC------GCACGACGACGGCGACAACGACGAC--AC |
| droVir3 | scaffold_12963:15419953-15420067 - | --------------CTGCG--CCGTCTTTTACCGTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----GGTTATCCCACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGGCCAGACGTATT--CATC------TT-------------------AAGC--TT |
| droMoj3 | scaffold_6500:18399386-18399492 - | -----------------------GTCTTTTACCGTGCGCGCTAATCAGTGACCGGGGCTGGTTTC-----GATTATCTCACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGGCTAGACGCATT--CAAC------GCTC-----------------T-----GC |
| droGri2 | scaffold_15126:5008400-5008494 + | -----------------------GTCGTTTACCGCGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----GATTATCCCACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGGCCAGACGCAT------------------------------------------ |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:7425780-7425907 + | TGATGTTCCCCCGACTGTA--AAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----TTATAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGG-CAGACATATAT----CTCCATCTT-------------------CACC--AT |
| droSim1 | chr2L:7217350-7217477 + | TGATGTTCCCCCGACTGAA--AAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----TTATAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGG-CAGACATATAT----CTCCATCTC-------------------CGCC--AT |
| droSec1 | super_3:2941240-2941367 + | TGATGTTCCCCCGACTGAA--AAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTTGTTTT-----TTATAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGG-CAGACATATAT----CTCCATCTC-------------------CGTC--AT |
| droYak2 | chr2L:5473766-5473897 - | TGATGTTCCCCCGACTGAACAGAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----TTATAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGC-CAGACATATAT----CTCCAACTC-------------------CACGATAT |
| droEre2 | scaffold_4929:16344506-16344633 + | TGATGTTCCCCCGACTGAA--AAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----TTATAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGC-CAGACATATAT----CTCCATCTC-------------------CACC--AT |
| droAna3 | scaffold_12916:15333987-15334092 + | -------------ACTCTC--CAGTCTCCTACCTTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----TGCA-----ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGG-CAGACATATAT----C---------------------------CGCC--AT |
| dp4 | chr4_group1:4175726-4175840 + | CCACCTTC-------TTGA--AAGTCTCCTACCATGCGCGCTAATCAGAGACCGGGGCTGGTTTT-----TTCACT---GCAGTCAGGTACCTGAAGTAGCGCGCGTGGTGC-TAGACACGAA--CATC--TGT----------------------------GT |
| droPer1 | super_5:1086449-1086563 - | CCACCTTC-------TTGA--AAGTCTCCTACCATGCGCGCTAATCAGAGACCGGGGCTGGTTTT-----TTCACT---GCAGTCAGGTACCTGAAGTAGCGCGCGTGGTGC-TAGACACGAA--CATC--TGT----------------------------GT |
| droWil1 | scaffold_180772:5257564-5257692 - | -----------------------GTCTCTTACCTTGCGCGCTAATCAGTGACCAGGGCTGAGTAATTTTATTCAAT---ACAGTCAGGTACCTGAAGTAGCGCGCGTGGTTT-TAGACCAAGATTCATC------GCACGACGACGGCGACAACGACGAC--AC |
| droVir3 | scaffold_12963:15419953-15420067 - | --------------CTGCG--CCGTCTTTTACCGTGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----GGTTATCCCACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGGCCAGACGTATT--CATC------TT-------------------AAGC--TT |
| droMoj3 | scaffold_6500:18399386-18399492 - | -----------------------GTCTTTTACCGTGCGCGCTAATCAGTGACCGGGGCTGGTTTC-----GATTATCTCACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGGCTAGACGCATT--CAAC------GCTC-----------------T-----GC |
| droGri2 | scaffold_15126:5008400-5008494 + | -----------------------GTCGTTTACCGCGCGCGCTAATCAGTGACCGGGGCTGGTTTT-----GATTATCCCACAGTCAGGTACCTGAAGTAGCGCGCGTGGTGGCCAGACGCAT------------------------------------------ |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
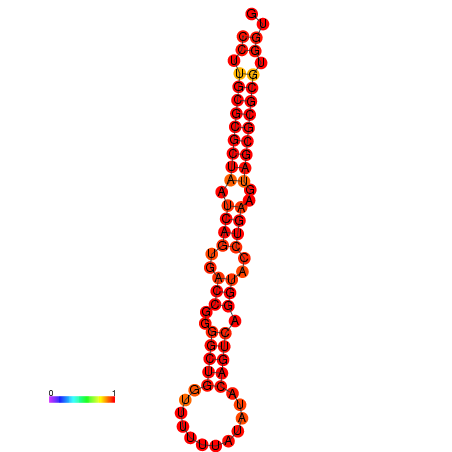
| dG=-33.0, p-value=0.009901 |
|---|
 |
| dG=-33.0, p-value=0.009901 |
|---|
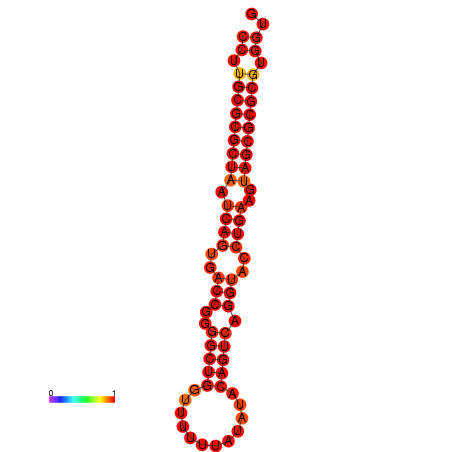 |
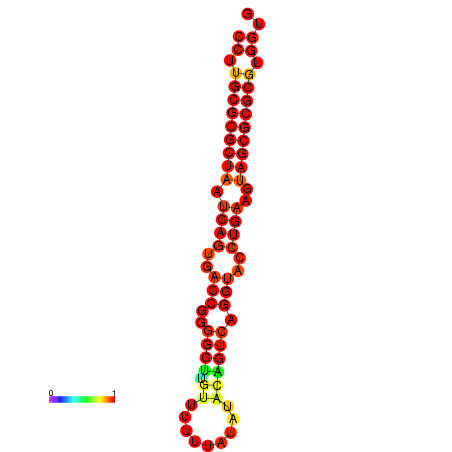
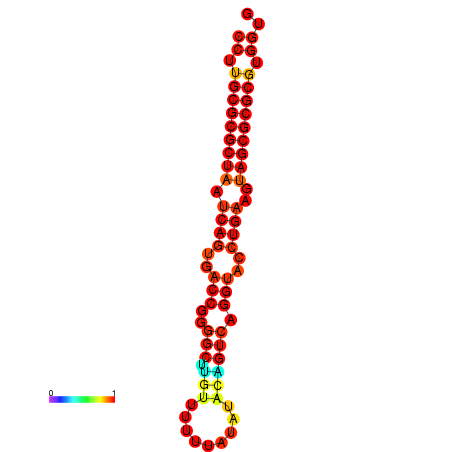
| dG=-30.1, p-value=0.009901 | dG=-30.1, p-value=0.009901 | dG=-29.5, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-33.0, p-value=0.009901 |
|---|
 |
| dG=-33.0, p-value=0.009901 |
|---|
 |
| dG=-33.2, p-value=0.009901 | dG=-32.2, p-value=0.009901 |
|---|---|
 |
 |
| dG=-37.9, p-value=0.009901 |
|---|
 |
| dG=-37.9, p-value=0.009901 |
|---|
 |
| dG=-31.2, p-value=0.009901 | dG=-31.2, p-value=0.009901 | dG=-31.2, p-value=0.009901 | dG=-30.9, p-value=0.009901 | dG=-30.9, p-value=0.009901 |
|---|---|---|---|---|
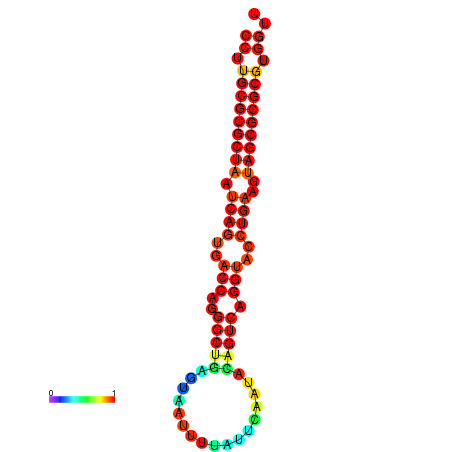 |
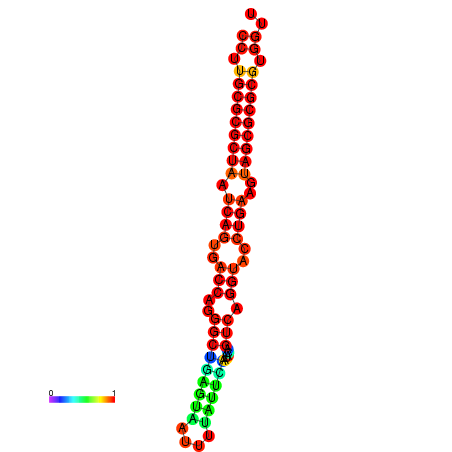 |
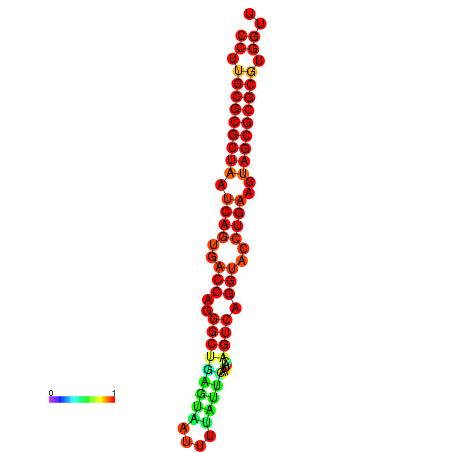 |
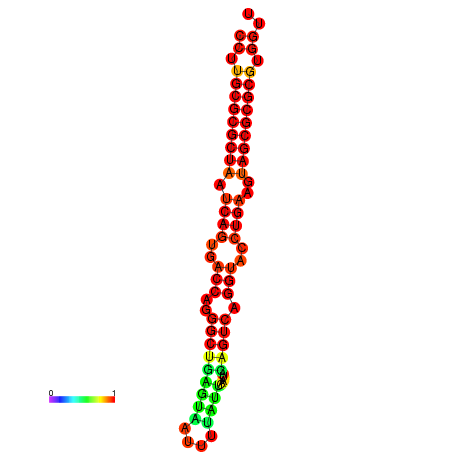 |
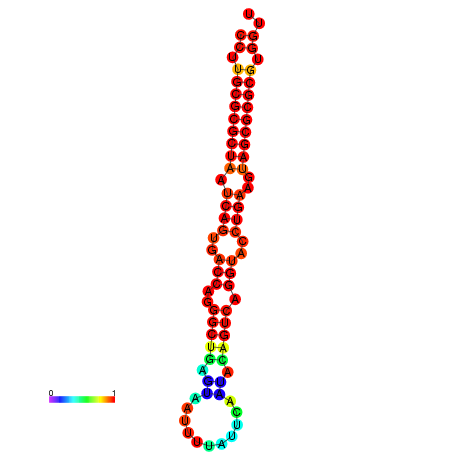 |
| dG=-35.0, p-value=0.009901 | dG=-34.8, p-value=0.009901 | dG=-34.0, p-value=0.009901 |
|---|---|---|
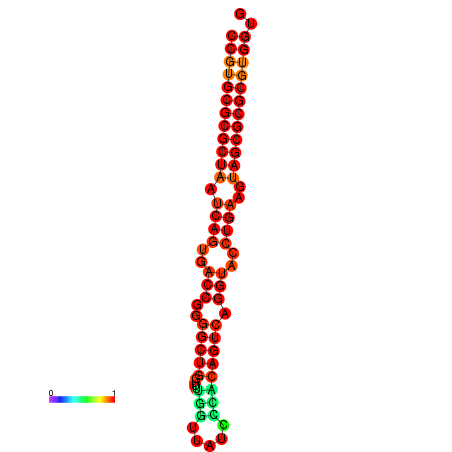 |
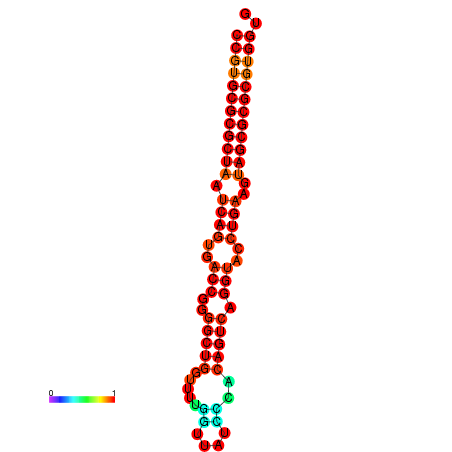 |
 |
| dG=-34.0, p-value=0.009901 |
|---|
 |
| dG=-38.8, p-value=0.009901 |
|---|
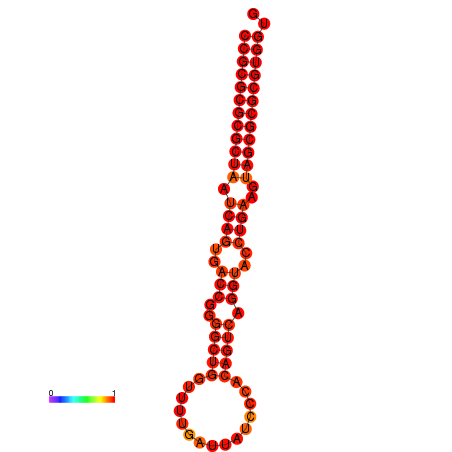 |
Generated: 03/07/2013 at 05:00 PM