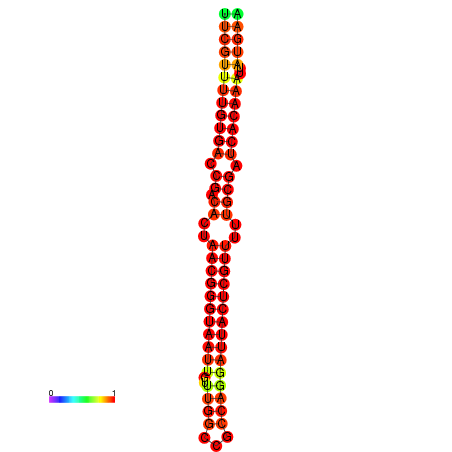
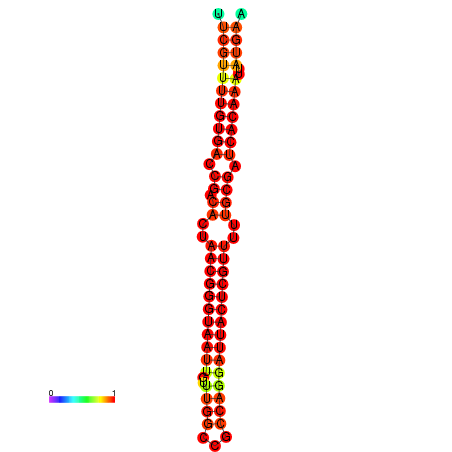
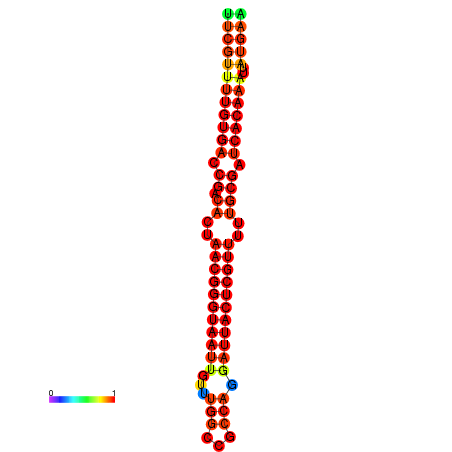
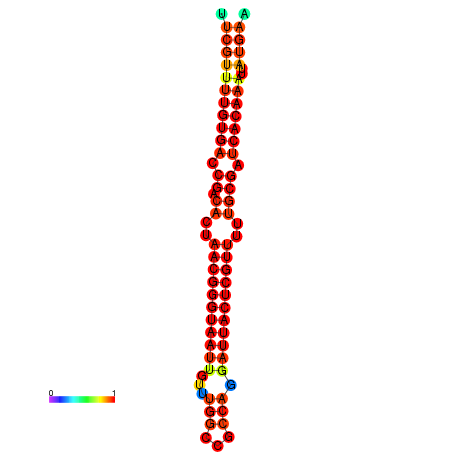
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:11650003-11650132 + | AG-T----TTTGGTGAC--------GAATCC---------TGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTG--------------------TT------------------TGGCCG-CCAGGAT--TACTCGTTTTTGCGATCACAAATTATGAAATTGCAGCAAAA-CT---------CAACGA--------AATTG |
| droSim1 | zdo16e12.g1:275-405 + | AG-T----TTTAGTGAC--------AAATCC---------TGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTG--------------------TT------------------TGGCCA-CCAGGAT--TACTCGTTTTTGCGATCACAAATCATGAAATTGCAGCAAAA-TG---------CAACAA--------AAATG |
| droSec1 | super_0:3837012-3837141 + | AG-T----TTTAGTGAC--------AAATCC---------TGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTG--------------------TT------------------TGATCA-CCAGGAT--TACTCGTTTTTGCGATCACAAATCATGAAATTGCAGCAAAA-CG---------CAACAA--------ATATA |
| droYak2 | chr3L:11678621-11678750 + | TG-A----TTTTATGCC--------GAATCC---------TGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTG--------------------TT------------------TGGTCG-CCAGGAT--TACTCGTTTTTGCGATCACAAATCATGAAATTGCAGCAAAA-CG---------CAACAA--------AGTCG |
| droEre2 | scaffold_4784:11632283-11632412 + | TG-A----TTTTATGGC--------CAATCC---------TGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTG--------------------TT------------------TGGCCG-CCAGGAT--TACTCGTTTTTGCGATCACAAATCATGAAATTGCAGCAAAA-CG---------CAACGA--------AGTCG |
| droAna3 | scaffold_13337:5918600-5918728 + | GC-C----TTTTTTTAA--------AAAACA-------GAAATGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTATTTT--------------------AC------------------AGGACATACAGGAT--TACTCGTTTTTGCGATCACAAATCCTGAAATTGCAGCAAAA-AA---------CAGCAA--------A---- |
| dp4 | chrXR_group6:10040179-10040309 - | CC-TGGCACCTGGAACC--------TGGTCC---------CGTGCTGCAGTTTCATTTTGTGACCGACACTAACGGGTAATGG---------------------C------------------TGGCCG--CAGGAC--TACTCGTTTTTGCGATCACA-ATACTGAAATTGCAGCAACG-AG---------CAGCGA--------AACTG |
| droPer1 | super_61:320241-320372 - | ACCTGGCACCTGGGACC--------TGGTCC---------CGTGCTGCAGTTTCATTTTGTGACCGACACTAACGGGTAATGG---------------------C------------------TGGCCG--CAGGAC--TACTCGTTTTTGCGATCACA-ATACTGAAATTGCAGCAACG-AG---------CAGCGA--------AACTG |
| droWil1 | scaffold_180949:2705123-2705289 - | TT-T-------TGTGAT--------TATTTT---TAATATCCTGTTGCAGTTTCATTTTGTGACCGACACTAACGGGTAGTAACGGGTTAACAAAGTCGTTTTTC------------------TGGCCT------TT--TACTCGTTTTTGCGATCACAAATTCTGAAATTGCAGCAAAAGATCAACGTATACAACAAACACATAAAATGA |
| droVir3 | scaffold_13049:23860698-23860839 + | AA-T----CCCAATGTC--------GCATCCTGT-------TTGTTGCAGTTTCGGTTTGTGACCGACACTAACGGGTAGTTGGG----------------------GCA-CCTCGAATAC-G--------CTGTTT--TACTCGTTTTTGCGATCACAAATTCTGAAATTGCAACGAAA-AG---------CAACAA-----GAAAATTT |
| droMoj3 | scaffold_6680:18485462-18485604 + | GA-A----TCTGTTGTCAAGT----GTATCC---------TGTGTTGCAGTTTCGGTTTGTGACCGACACTAACGGGTAGTTGAG----------------------GCA-TCTCGTATTC-G--------CTGTTTTATACTCGTTTTTGCGATCACAAATTCTGAAATTGCAACGAAA-CG---------CAACGA--------AAATG |
| droGri2 | scaffold_15110:13805592-13805748 - | AA-AGTGAAATAGTGCAACGTATACGAGTCCTGT-----GCGTGTTGCAGTTTCGGTTTGTGACCGACACTAACGGGTAGTTA--------------------AAGGGCATCCTCGCATTCTG--------CTGGCT--TACTCGTTTTTGCGATCACAAATTCTGAAATTGCAACGAAA-AT---------CAACAA--------AACCA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:11650003-11650132 + | AG-T----TTTGGTGAC--------GAATCC---------TGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTG--------------------TT------------------TGGCCG-CCAGGAT--TACTCGTTTTTGCGATCACAAATTATGAAATTGCAGCAAAA-CT---------CAACGA--------AATTG |
| droSim1 | zdo16e12.g1:275-405 + | AG-T----TTTAGTGAC--------AAATCC---------TGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTG--------------------TT------------------TGGCCA-CCAGGAT--TACTCGTTTTTGCGATCACAAATCATGAAATTGCAGCAAAA-TG---------CAACAA--------AAATG |
| droSec1 | super_0:3837012-3837141 + | AG-T----TTTAGTGAC--------AAATCC---------TGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTG--------------------TT------------------TGATCA-CCAGGAT--TACTCGTTTTTGCGATCACAAATCATGAAATTGCAGCAAAA-CG---------CAACAA--------ATATA |
| droYak2 | chr3L:11678621-11678750 + | TG-A----TTTTATGCC--------GAATCC---------TGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTG--------------------TT------------------TGGTCG-CCAGGAT--TACTCGTTTTTGCGATCACAAATCATGAAATTGCAGCAAAA-CG---------CAACAA--------AGTCG |
| droEre2 | scaffold_4784:11632283-11632412 + | TG-A----TTTTATGGC--------CAATCC---------TGTGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTAATTG--------------------TT------------------TGGCCG-CCAGGAT--TACTCGTTTTTGCGATCACAAATCATGAAATTGCAGCAAAA-CG---------CAACGA--------AGTCG |
| droAna3 | scaffold_13337:5918600-5918728 + | GC-C----TTTTTTTAA--------AAAACA-------GAAATGTTGCAGTTTCGTTTTGTGACCGACACTAACGGGTATTTT--------------------AC------------------AGGACATACAGGAT--TACTCGTTTTTGCGATCACAAATCCTGAAATTGCAGCAAAA-AA---------CAGCAA--------A---- |
| dp4 | chrXR_group6:10040179-10040309 - | CC-TGGCACCTGGAACC--------TGGTCC---------CGTGCTGCAGTTTCATTTTGTGACCGACACTAACGGGTAATGG---------------------C------------------TGGCCG--CAGGAC--TACTCGTTTTTGCGATCACA-ATACTGAAATTGCAGCAACG-AG---------CAGCGA--------AACTG |
| droPer1 | super_61:320241-320372 - | ACCTGGCACCTGGGACC--------TGGTCC---------CGTGCTGCAGTTTCATTTTGTGACCGACACTAACGGGTAATGG---------------------C------------------TGGCCG--CAGGAC--TACTCGTTTTTGCGATCACA-ATACTGAAATTGCAGCAACG-AG---------CAGCGA--------AACTG |
| droWil1 | scaffold_180949:2705123-2705289 - | TT-T-------TGTGAT--------TATTTT---TAATATCCTGTTGCAGTTTCATTTTGTGACCGACACTAACGGGTAGTAACGGGTTAACAAAGTCGTTTTTC------------------TGGCCT------TT--TACTCGTTTTTGCGATCACAAATTCTGAAATTGCAGCAAAAGATCAACGTATACAACAAACACATAAAATGA |
| droVir3 | scaffold_13049:23860698-23860839 + | AA-T----CCCAATGTC--------GCATCCTGT-------TTGTTGCAGTTTCGGTTTGTGACCGACACTAACGGGTAGTTGGG----------------------GCA-CCTCGAATAC-G--------CTGTTT--TACTCGTTTTTGCGATCACAAATTCTGAAATTGCAACGAAA-AG---------CAACAA-----GAAAATTT |
| droMoj3 | scaffold_6680:18485462-18485604 + | GA-A----TCTGTTGTCAAGT----GTATCC---------TGTGTTGCAGTTTCGGTTTGTGACCGACACTAACGGGTAGTTGAG----------------------GCA-TCTCGTATTC-G--------CTGTTTTATACTCGTTTTTGCGATCACAAATTCTGAAATTGCAACGAAA-CG---------CAACGA--------AAATG |
| droGri2 | scaffold_15110:13805592-13805748 - | AA-AGTGAAATAGTGCAACGTATACGAGTCCTGT-----GCGTGTTGCAGTTTCGGTTTGTGACCGACACTAACGGGTAGTTA--------------------AAGGGCATCCTCGCATTCTG--------CTGGCT--TACTCGTTTTTGCGATCACAAATTCTGAAATTGCAACGAAA-AT---------CAACAA--------AACCA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-26.3, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
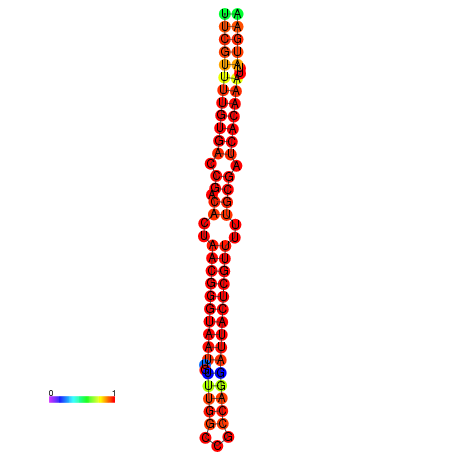
| dG=-26.3, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|---|---|
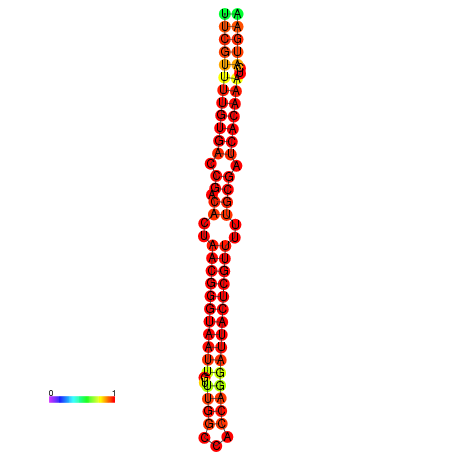 |
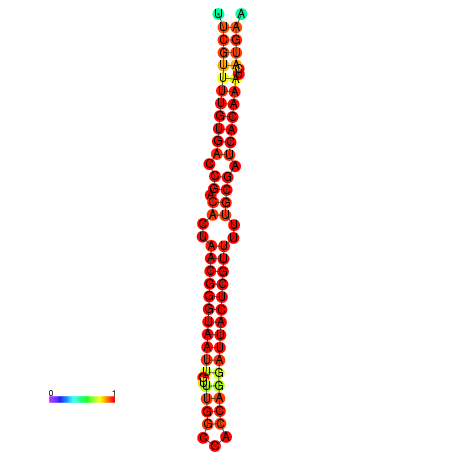 |
 |
 |
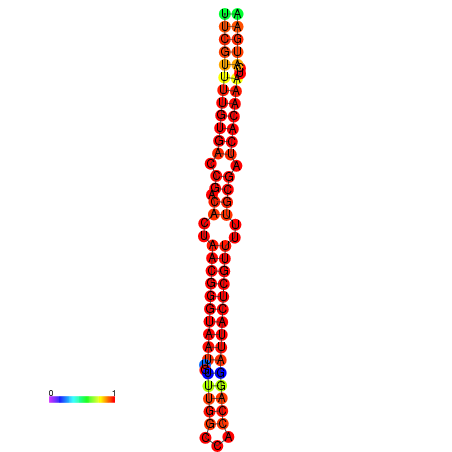 |
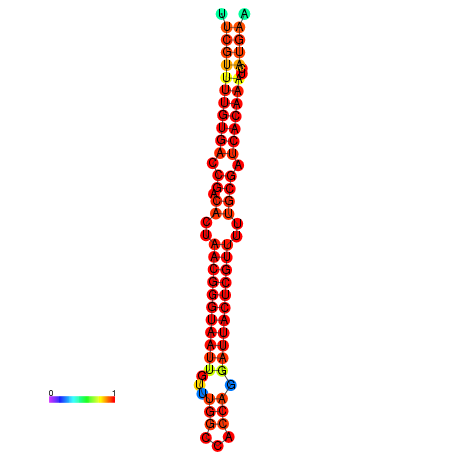
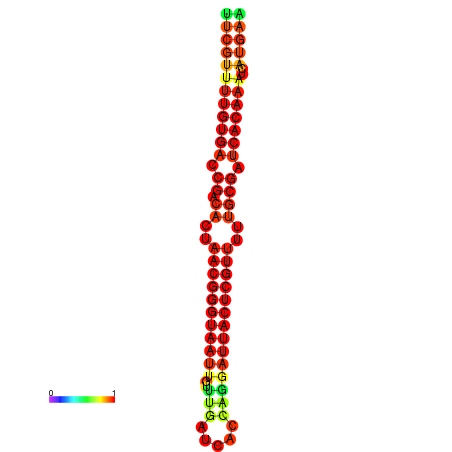
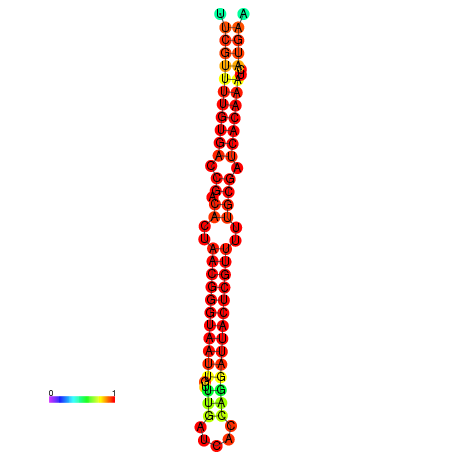
| dG=-24.2, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.7, p-value=0.009901 | dG=-23.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
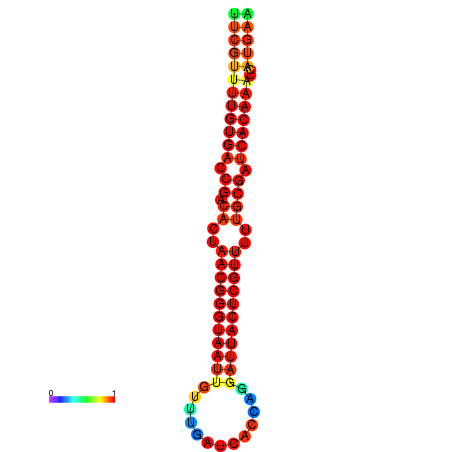 |
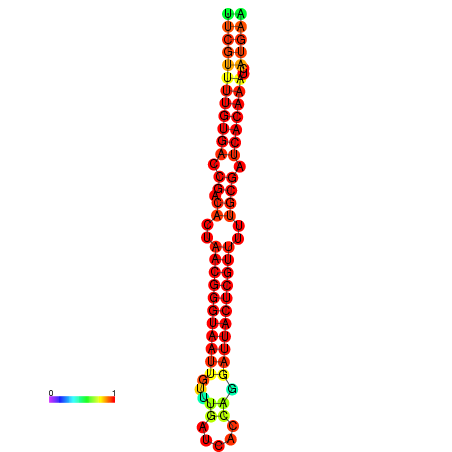 |
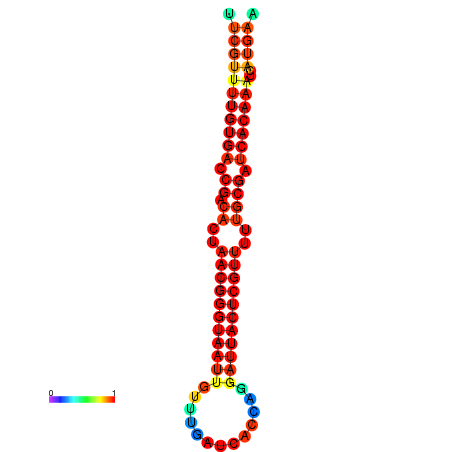 |
| dG=-26.3, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|---|---|
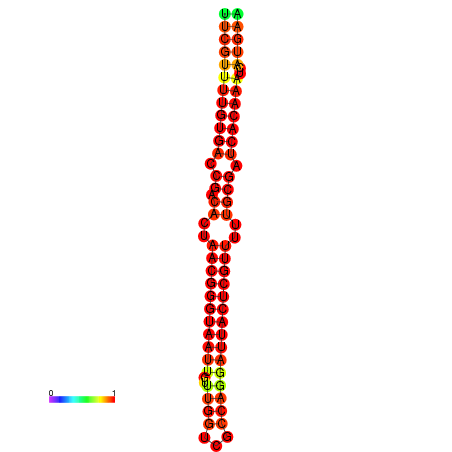 |
 |
 |
 |
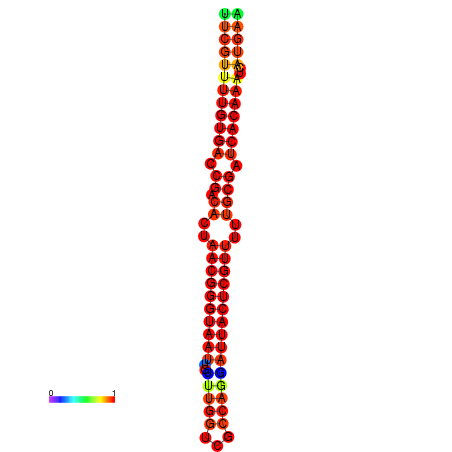 |
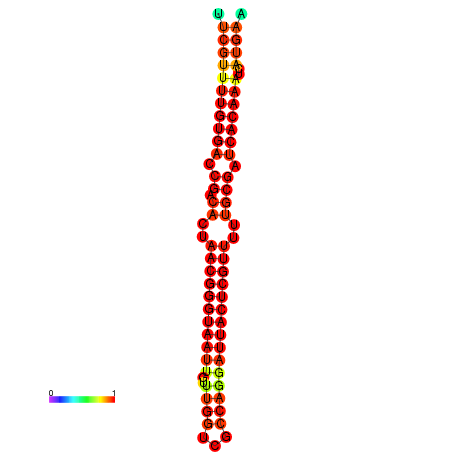
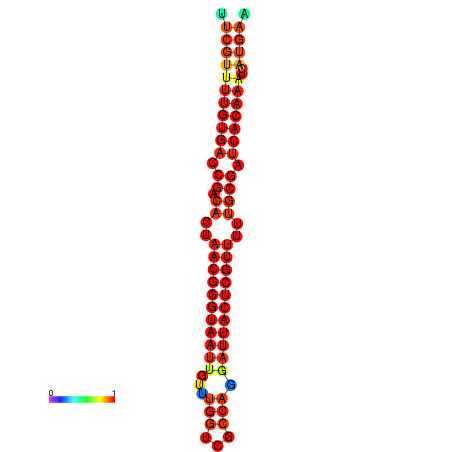
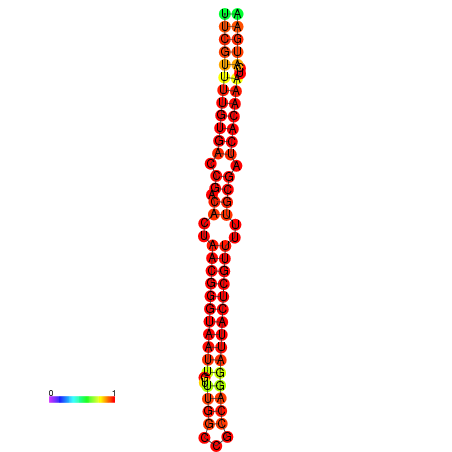
| dG=-26.3, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
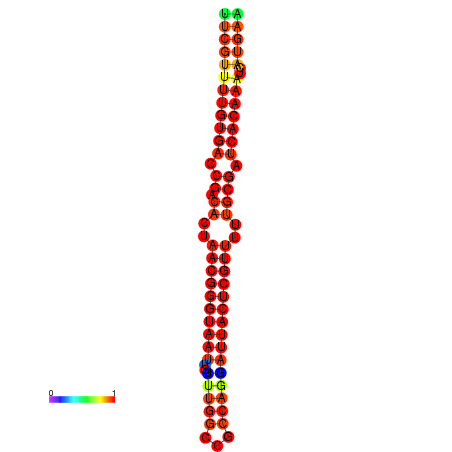 |
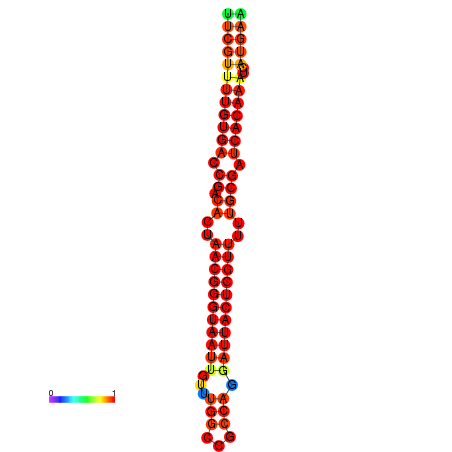
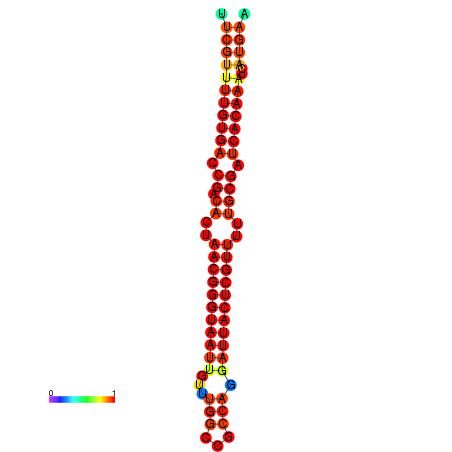
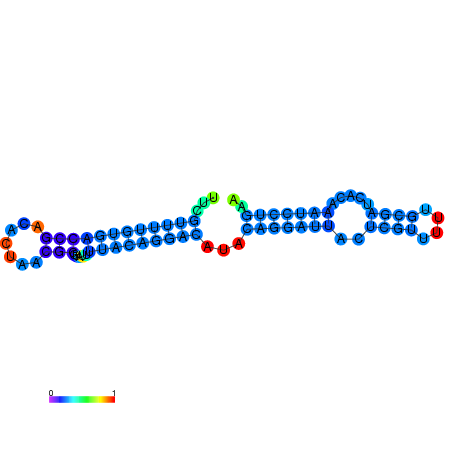
| dG=-19.3, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.1, p-value=0.009901 | dG=-18.9, p-value=0.009901 | dG=-18.8, p-value=0.009901 |
|---|---|---|---|---|
 |
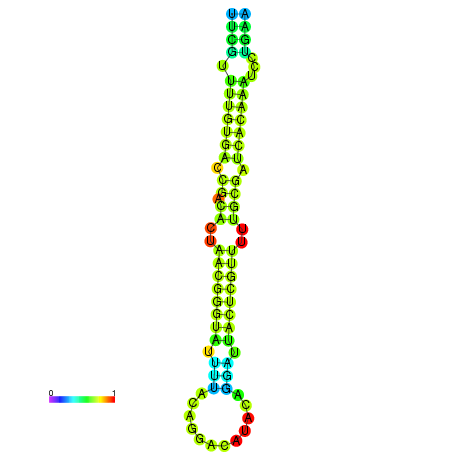 |
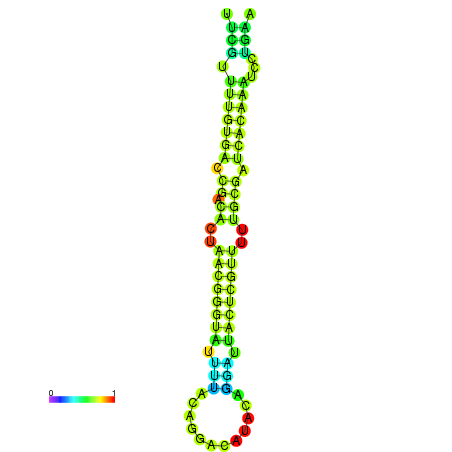 |
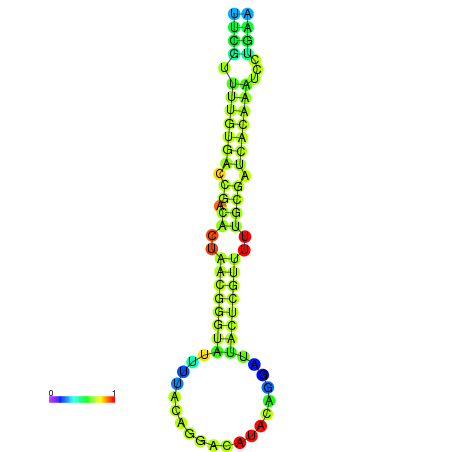 |
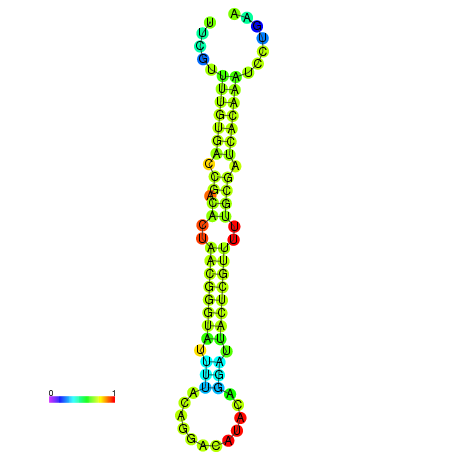 |
| dG=-23.1, p-value=0.009901 | dG=-22.9, p-value=0.009901 |
|---|---|
 |
 |
| dG=-23.1, p-value=0.009901 | dG=-22.9, p-value=0.009901 |
|---|---|
 |
 |
| dG=-25.6, p-value=0.009901 | dG=-25.4, p-value=0.009901 |
|---|---|
 |
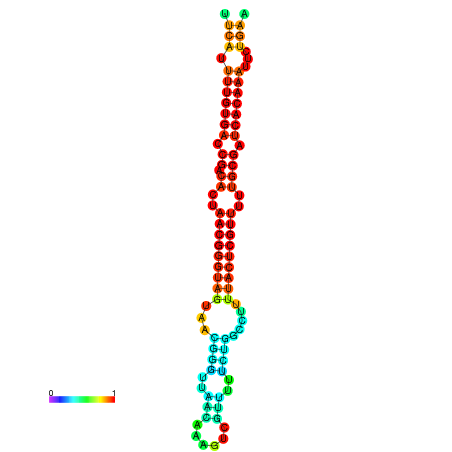 |
| dG=-25.9, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.3, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 |
|---|---|---|---|---|
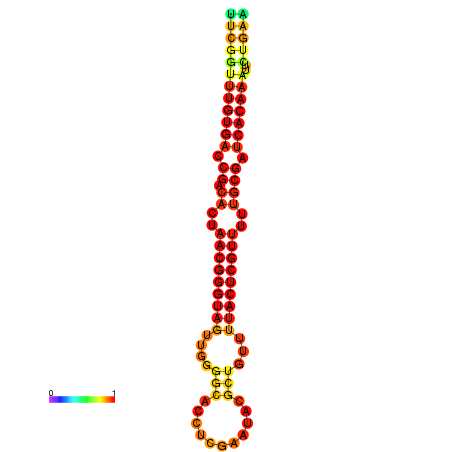 |
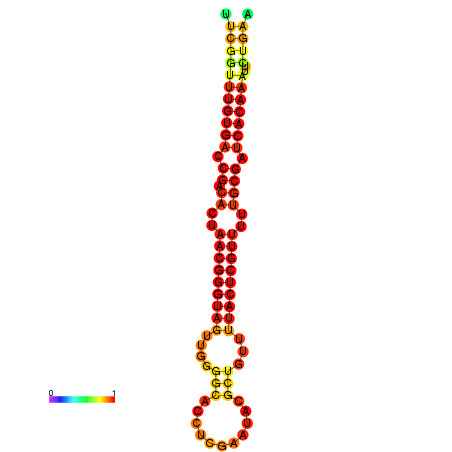 |
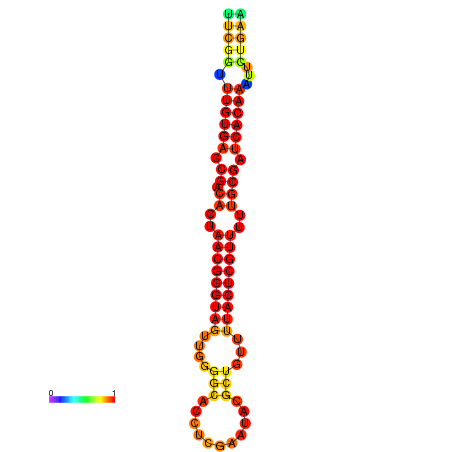 |
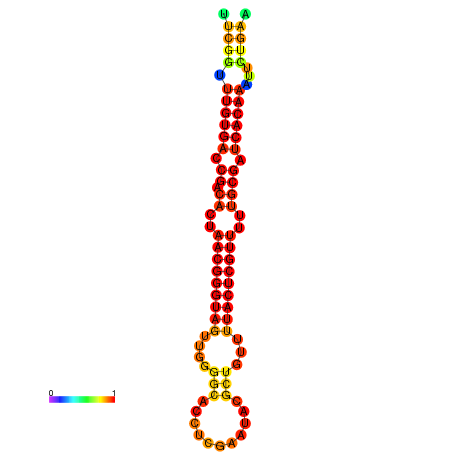 |
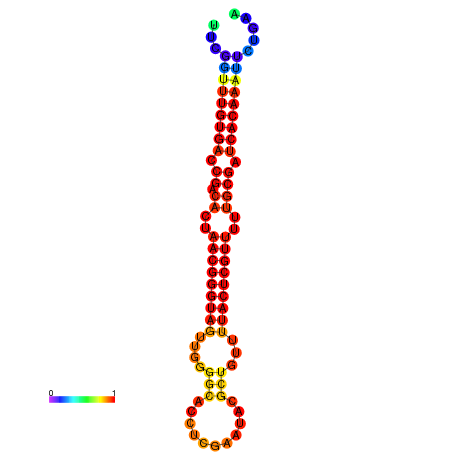 |
| dG=-27.0, p-value=0.009901 | dG=-26.8, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.1, p-value=0.009901 |
|---|---|---|---|---|
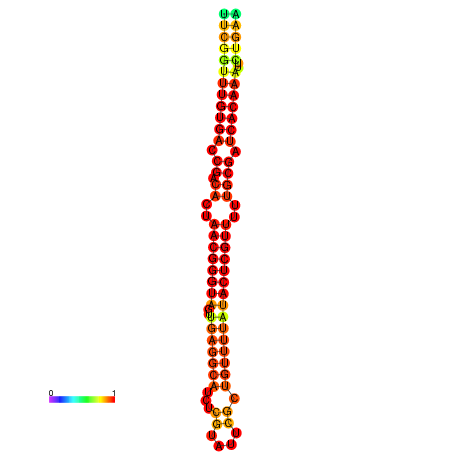 |
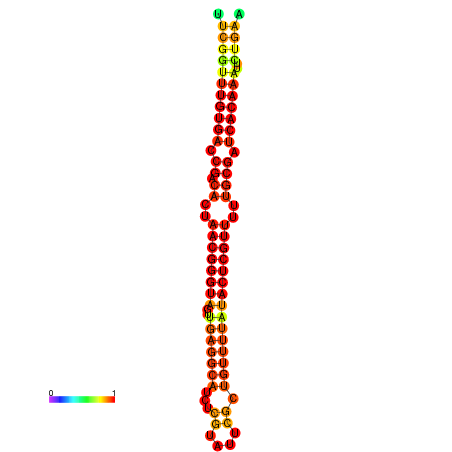 |
 |
 |
 |
| dG=-26.9, p-value=0.009901 | dG=-26.9, p-value=0.009901 | dG=-26.8, p-value=0.009901 | dG=-26.8, p-value=0.009901 | dG=-26.7, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
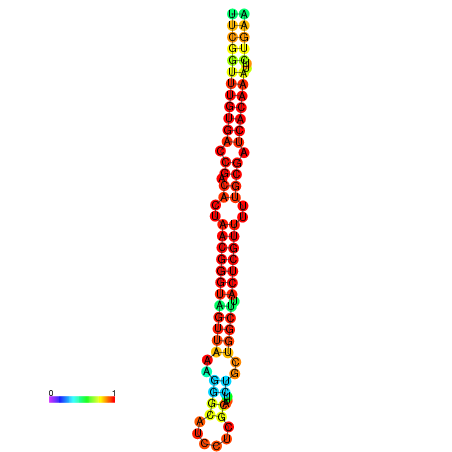 |
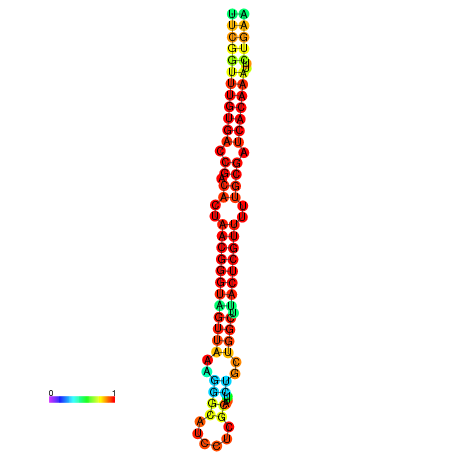 |
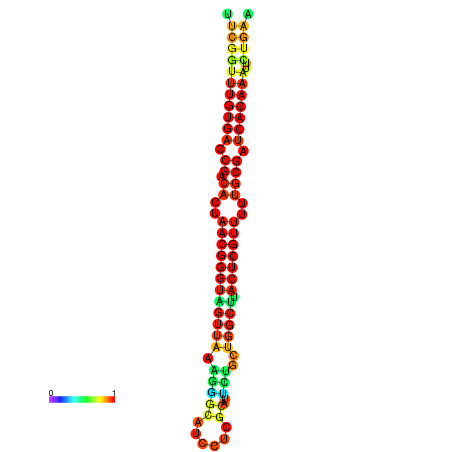 |
Generated: 03/07/2013 at 05:01 PM