| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:15836491-15836610 - | CTGTAGCTCTTCA-----------------------------A------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTGTACAACGATT------------TCT |
| droSim1 | chr3L:15181316-15181435 - | CTCTAGCTCTTCA-----------------------------A------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTGAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTATACAACGAAT------------TCT |
| droSec1 | super_0:7944386-7944505 - | CGCTAGCTCTTCA-----------------------------A------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTGAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTATACAACGATT------------TCT |
| droYak2 | chr3L_random:2539138-2539257 - | CGATAGCTCTTCA-----------------------------A------TCTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTTAATCAACG-GCTCTATACAACGATT------------TCT |
| droEre2 | scaffold_4784:18090156-18090270 - | CTG--GCGATTCA-----------------------------AACC---ATAGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGATTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTTAATCAACA-GCTCCATACAACG------------------ |
| droAna3 | scaffold_13337:11320710-11320858 + | TTGTGATTTTTGAATCAAATATTCTG--------------G----ACTCTCTGCTGTCTTTGAATCTTGGCACTGGGAGAATTCACAGTTGATTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTGAAATAATG-GCCTAAAACCTCAAAAAAGGATATTTATTAT |
| dp4 | chrXR_group6:3598033-3598148 - | CGATCGTTA-----------------------------------------TTGCTGGCATTGGCTCTTGGCACTGGGAGAATTCACAGTTGGTTTCCAT---ATATTCTGTGGTTCTGCGGGTGCC-AAAATCGAATGTTCG-GCTCGAAACAGCAGCT------------GAA |
| droPer1 | super_9:1894031-1894138 - | T--------------------------------------------------TGCTGGCATTGGCTCTTGGCACTGGGAGAATTCACAGTTGGTTTTCAT---ATATTCTGTGGTTCTGCGGGTGCCCAAAATCGAATGTTCG-GCTCGAAACAGCAGCT------------GAA |
| droWil1 | scaffold_180727:1452378-1452537 + | CTTAAATTCTAAG-AAAGATTTTCTGTGTGTGTTTGAACGAAAATAAAGTCTGCTCACAATGCATCTTGGCACTGGGAGAATTCACAGTTGATTCTTATTCGATATTCTGTGGTTCTGCGGGTGCC-AAAATCTAATGTTTGAGCTGTGATCATCGAAA------------AAG |
| droVir3 | scaffold_13049:24412507-24412633 - | TTGT-GTTCTTCT------------------------ACGGAAAGCAGAGCTACTGCCATTTCATCTTGGCACTGGGAGAATTCACAGTCGATTACATA---ATATTCTGTGGTTCTGCGGGTGCC-AAAATGTAATGTGTG-GACCAAGACAGC-----------------CT |
| droMoj3 | Unknown | AC-TAGT---------------------------------------------------------GATTGGCACTGGGAGAATTCACAGTTGATTATATA---ATATTCTGTGGTTCTGCGGGTGCC-AAAATATAATATTCG-GAATA------------------------AT |
| droGri2 | scaffold_15110:24319786-24319904 - | ATATTGT--GGAA-----------------------------A------GCTACTGCCATTACATCTTGGCACTGGGAGAATTCACAGTTGGTTATATA---ATATTCTGTGGTTCTGCGGGTGCC-AAACTGTAATGTGTG-GACATAGA-AACAACC------------TGA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:15836491-15836610 - | CTGTAGCTCTTCA-----------------------------A------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTGTACAACGATT------------TCT |
| droSim1 | chr3L:15181316-15181435 - | CTCTAGCTCTTCA-----------------------------A------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTGAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTATACAACGAAT------------TCT |
| droSec1 | super_0:7944386-7944505 - | CGCTAGCTCTTCA-----------------------------A------TTTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTGAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAACTTAAAAACCG-GCTTTATACAACGATT------------TCT |
| droYak2 | chr3L_random:2539138-2539257 - | CGATAGCTCTTCA-----------------------------A------TCTGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGACTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTTAATCAACG-GCTCTATACAACGATT------------TCT |
| droEre2 | scaffold_4784:18090156-18090270 - | CTG--GCGATTCA-----------------------------AACC---ATAGCTGACTTTGAGTCTTGGCACTGGGAGAATTCACAGTTGATTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTTAATCAACA-GCTCCATACAACG------------------ |
| droAna3 | scaffold_13337:11320710-11320858 + | TTGTGATTTTTGAATCAAATATTCTG--------------G----ACTCTCTGCTGTCTTTGAATCTTGGCACTGGGAGAATTCACAGTTGATTTTAT-----TATTCTGTGGTTCTGCGGGTGCC-AAAATTGAAATAATG-GCCTAAAACCTCAAAAAAGGATATTTATTAT |
| dp4 | chrXR_group6:3598033-3598148 - | CGATCGTTA-----------------------------------------TTGCTGGCATTGGCTCTTGGCACTGGGAGAATTCACAGTTGGTTTCCA---TATATTCTGTGGTTCTGCGGGTGCC-AAAATCGAATGTTCG-GCTCGAAACAGCAGCT------------GAA |
| droPer1 | super_9:1894031-1894138 - | T--------------------------------------------------TGCTGGCATTGGCTCTTGGCACTGGGAGAATTCACAGTTGGTTTTCA---TATATTCTGTGGTTCTGCGGGTGCCCAAAATCGAATGTTCG-GCTCGAAACAGCAGCT------------GAA |
| droWil1 | scaffold_180727:1452378-1452537 + | CTTAAATTCTAAG-AAAGATTTTCTGTGTGTGTTTGAACGAAAATAAAGTCTGCTCACAATGCATCTTGGCACTGGGAGAATTCACAGTTGATTCTTATTCGATATTCTGTGGTTCTGCGGGTGCC-AAAATCTAATGTTTGAGCTGTGATCATCGAAA------------AAG |
| droVir3 | scaffold_13049:24412507-24412633 - | TTGT-GTTCTTCT------------------------ACGGAAAGCAGAGCTACTGCCATTTCATCTTGGCACTGGGAGAATTCACAGTCGATTACAT---AATATTCTGTGGTTCTGCGGGTGCC-AAAATGTAATGTGTG-GACCAAGACAGC-----------------CT |
| droGri2 | scaffold_15110:24319786-24319904 - | ATATTGT--GGAA-----------------------------A------GCTACTGCCATTACATCTTGGCACTGGGAGAATTCACAGTTGGTTATAT---AATATTCTGTGGTTCTGCGGGTGCC-AAACTGTAATGTGTG-GACATAGA-AACAACC------------TGA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
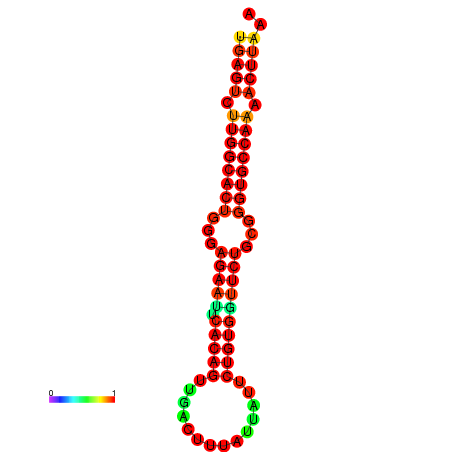
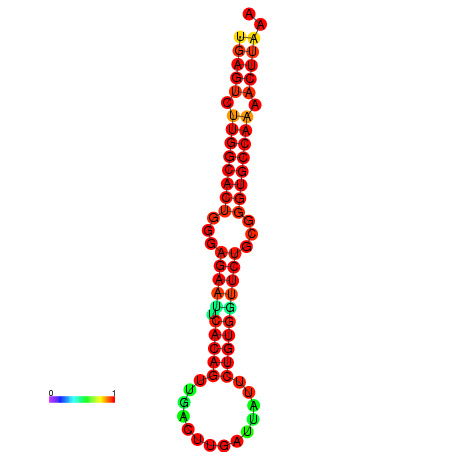
| dG=-27.3, p-value=0.009901 | dG=-27.3, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-27.1, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
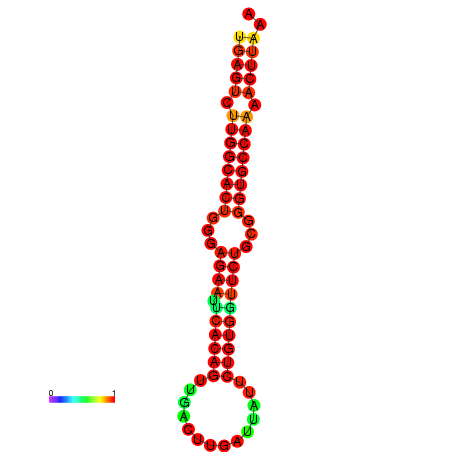
| dG=-27.3, p-value=0.009901 | dG=-27.3, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-27.1, p-value=0.009901 |
|---|---|---|---|
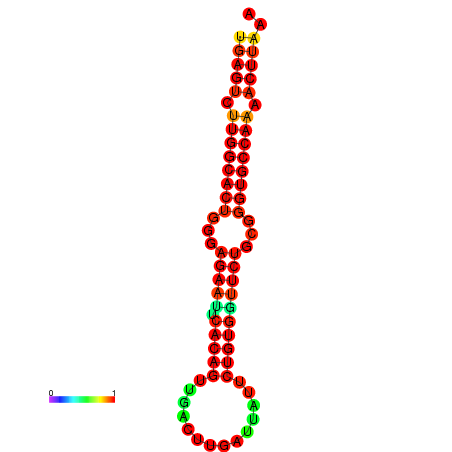 |
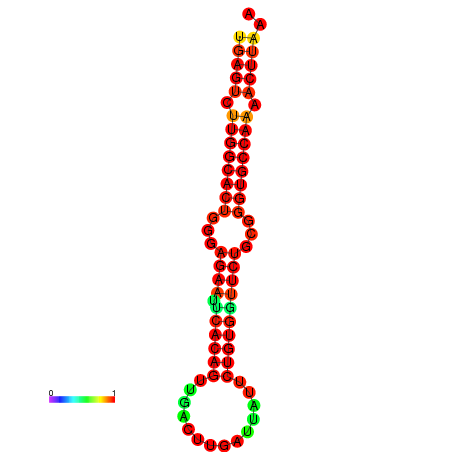 |
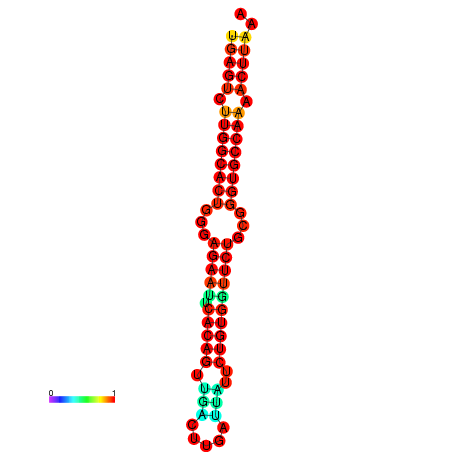 |
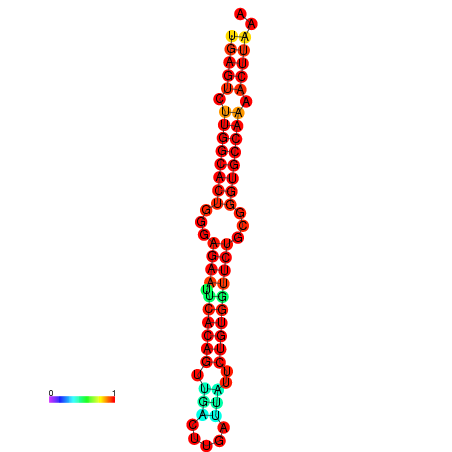 |
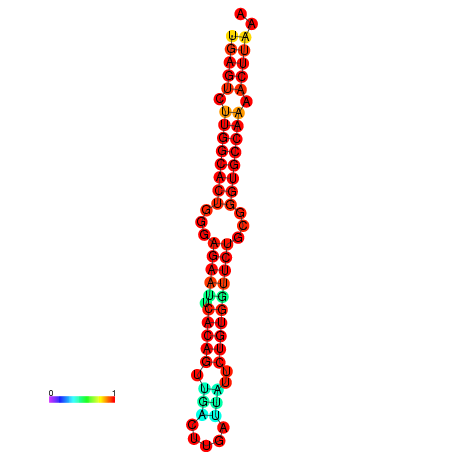
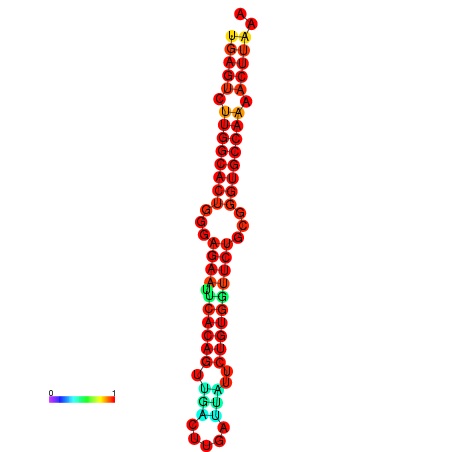
| dG=-27.3, p-value=0.009901 | dG=-27.3, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-27.1, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
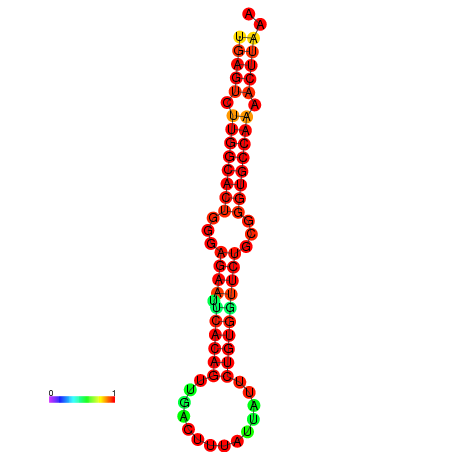
| dG=-25.0, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.8, p-value=0.009901 |
|---|---|---|---|
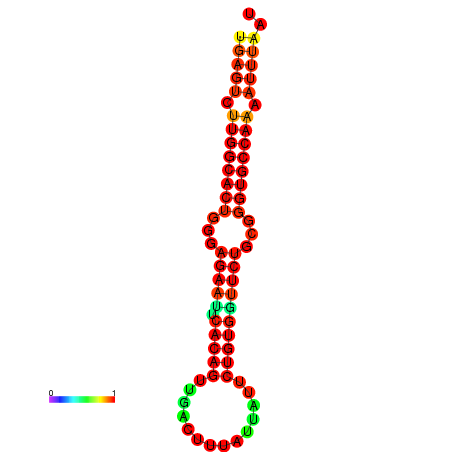 |
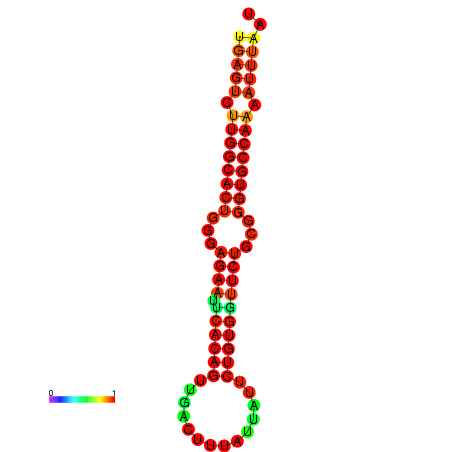 |
 |
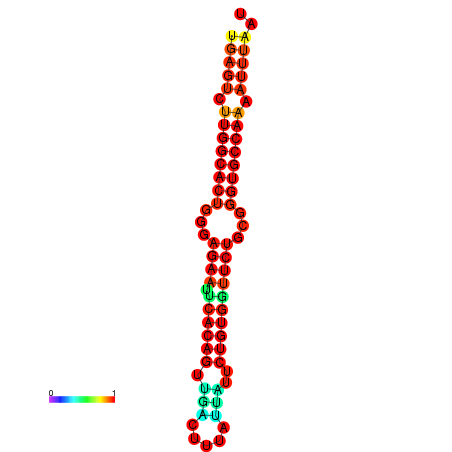 |
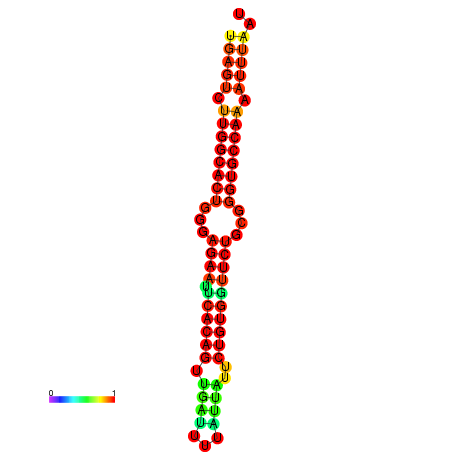
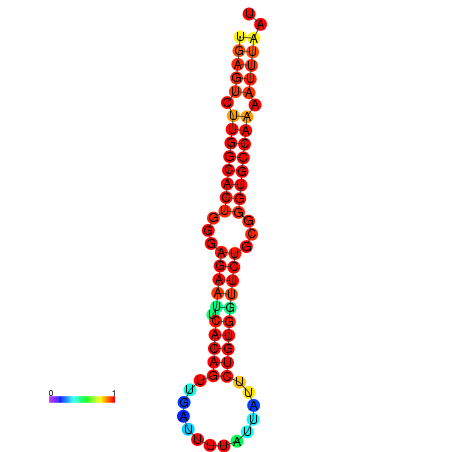
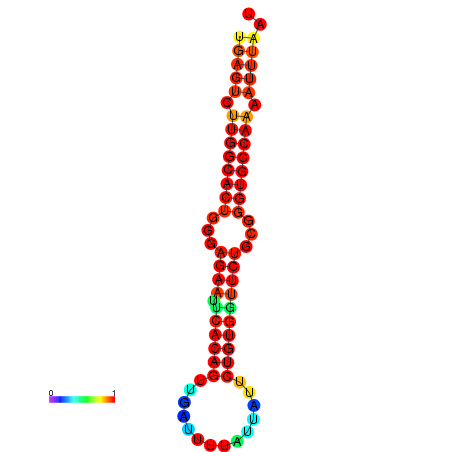
| dG=-25.6, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|---|---|---|
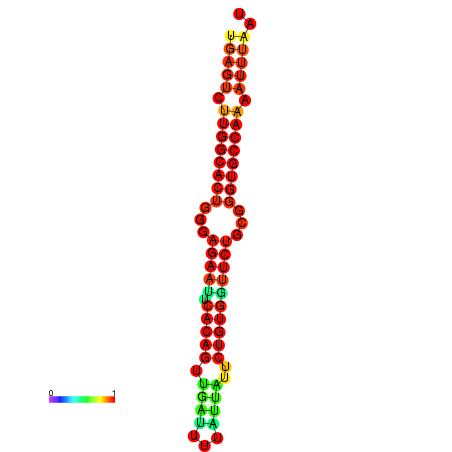 |
 |
 |
 |
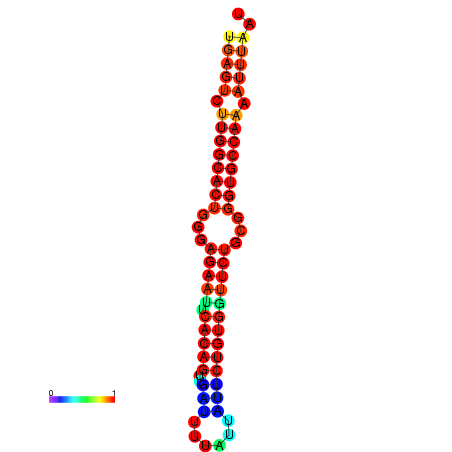 |
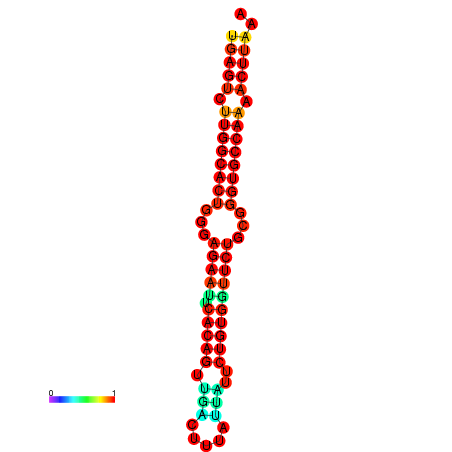
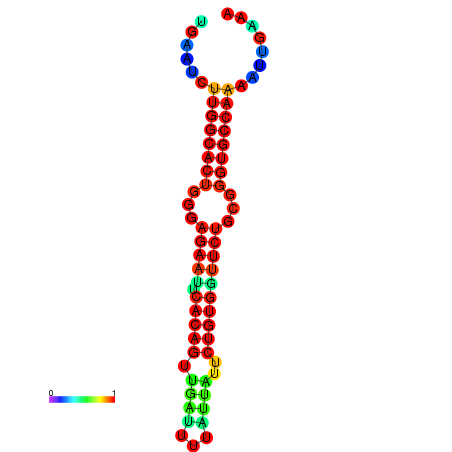
| dG=-23.4, p-value=0.009901 | dG=-23.4, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
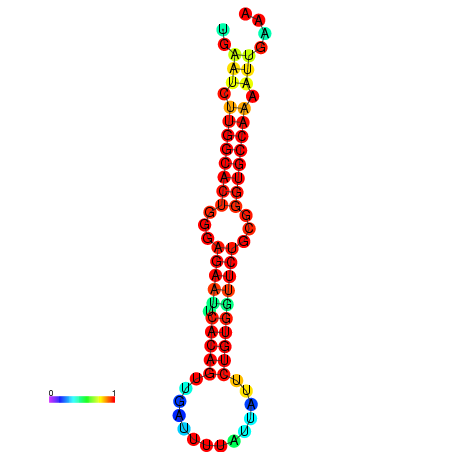 |
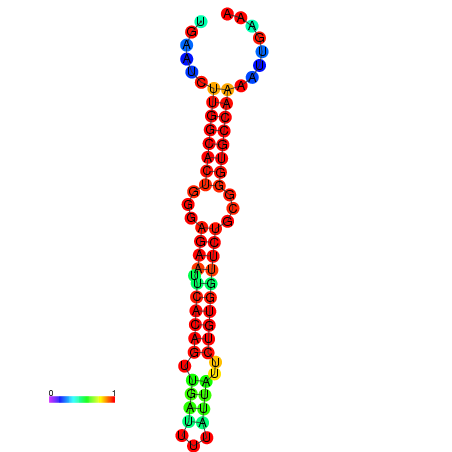
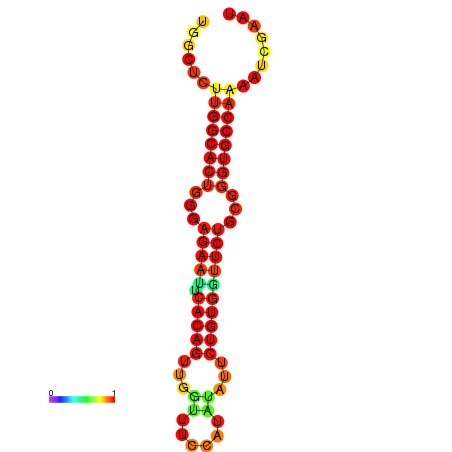
| dG=-23.5, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-22.6, p-value=0.009901 | dG=-22.6, p-value=0.009901 | dG=-22.5, p-value=0.009901 |
|---|---|---|---|---|
 |
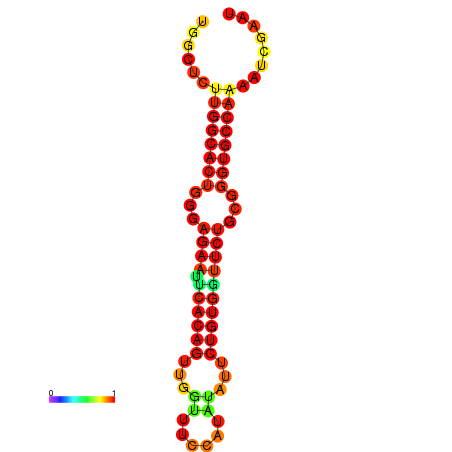 |
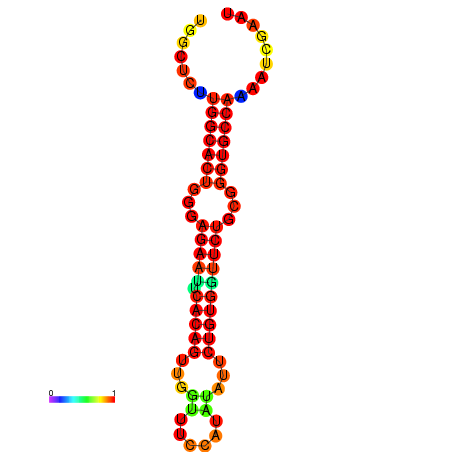 |
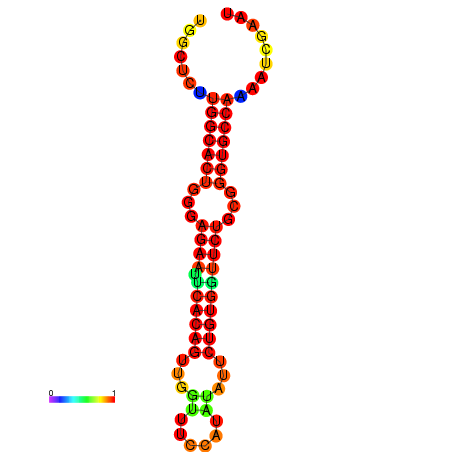 |
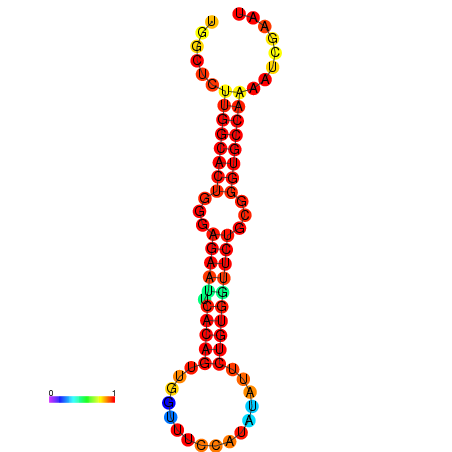 |
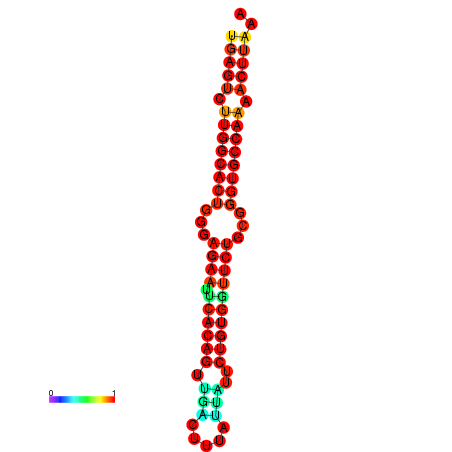
| dG=-21.3, p-value=0.009901 | dG=-21.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 |
|---|---|---|---|
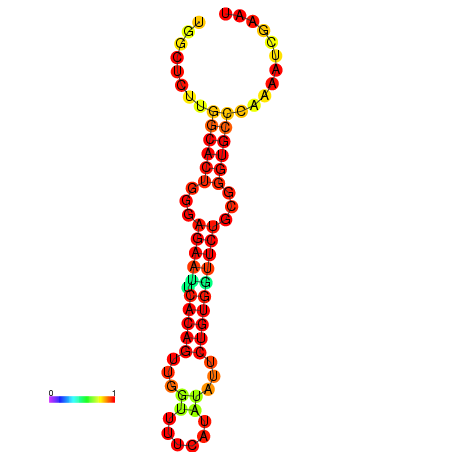 |
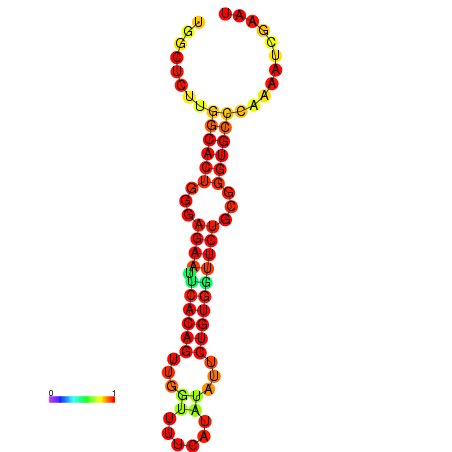 |
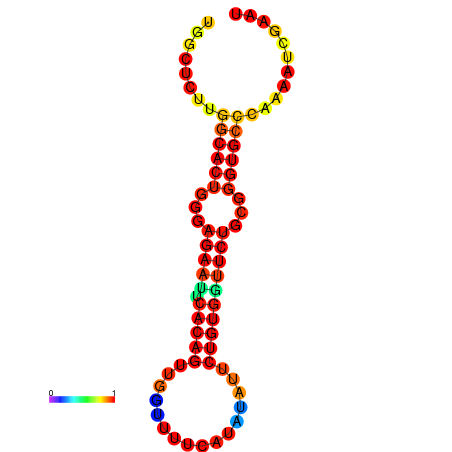 |
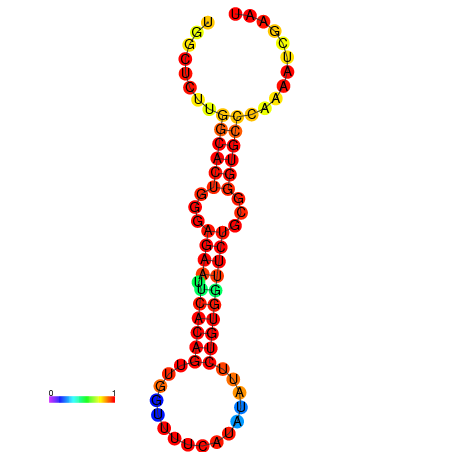 |
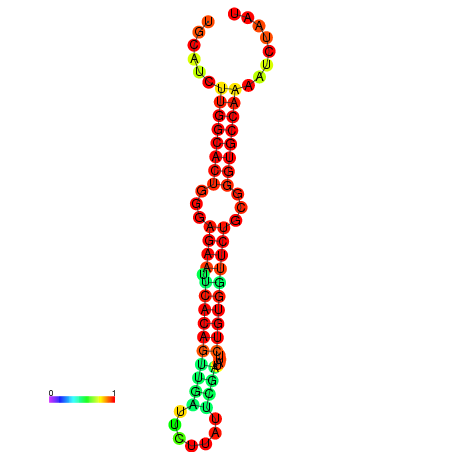
| dG=-24.1, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.3, p-value=0.009901 |
|---|---|---|---|---|
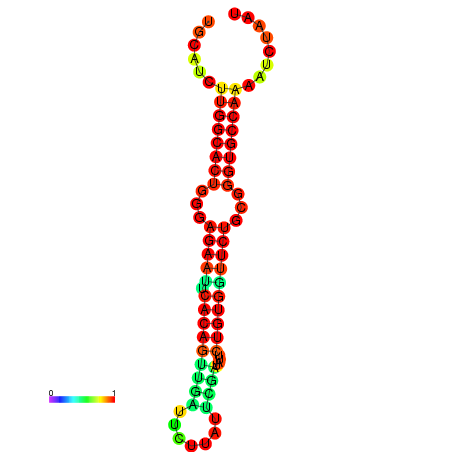 |
 |
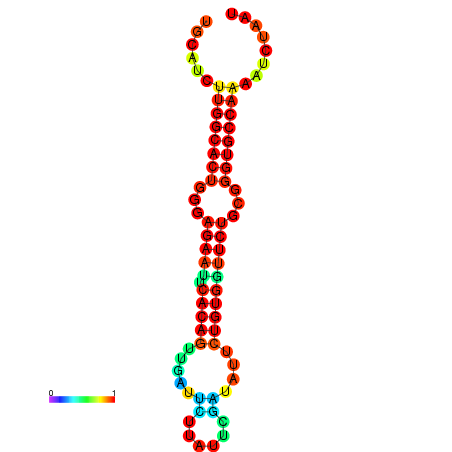 |
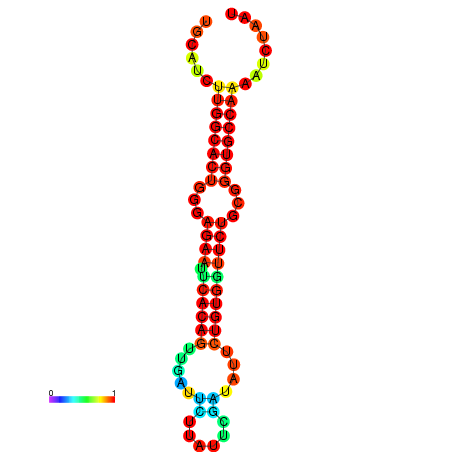 |
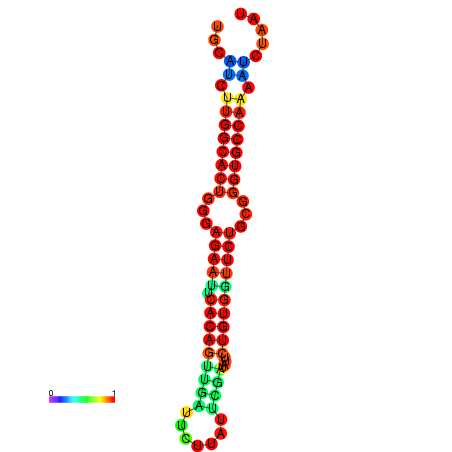 |
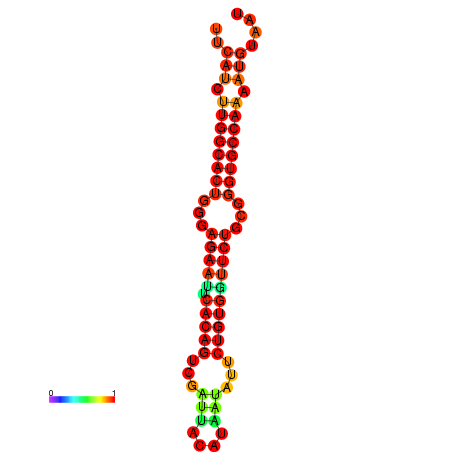
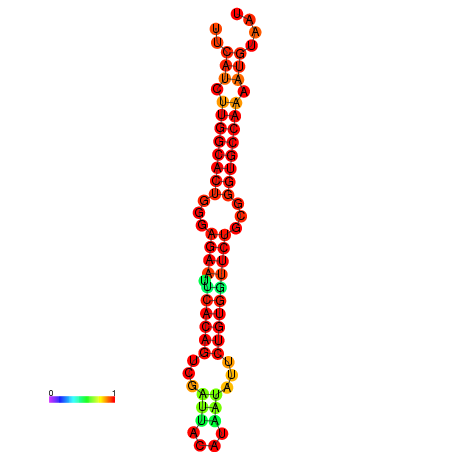
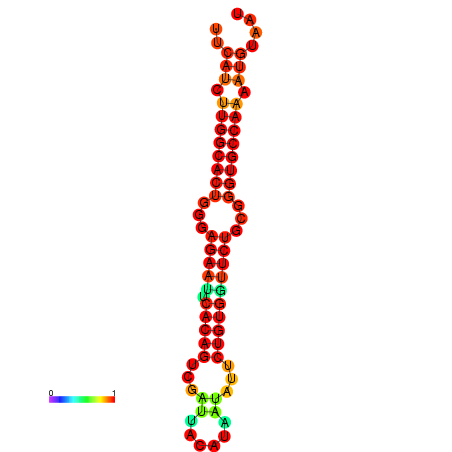
| dG=-25.6, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
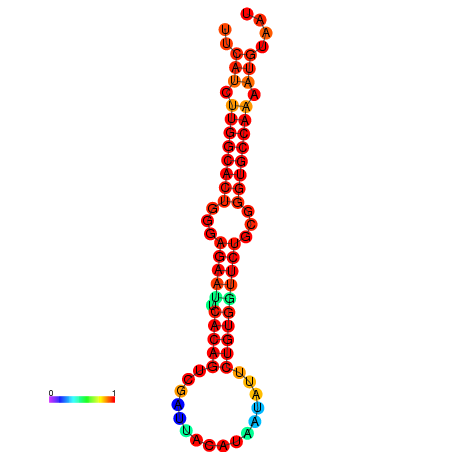 |
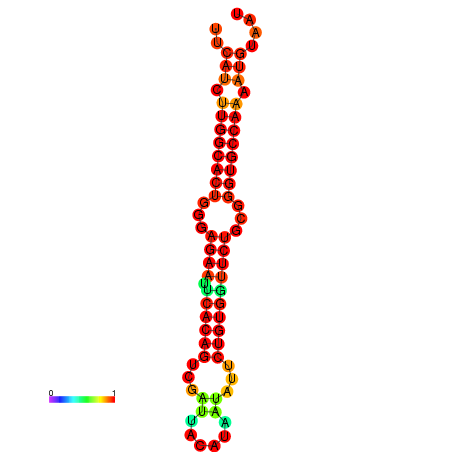 |
| dG=-23.6, p-value=0.009901 | dG=-23.6, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|---|
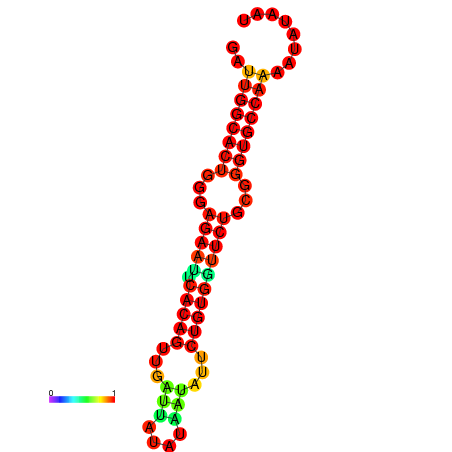 |
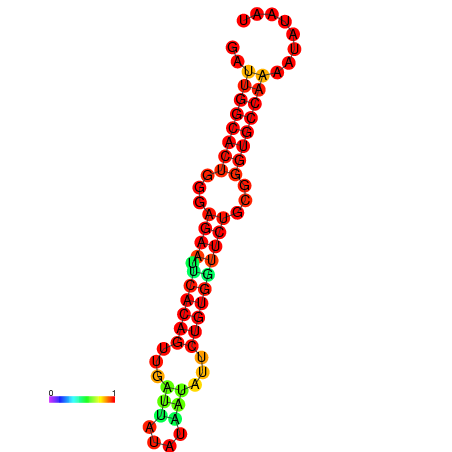 |
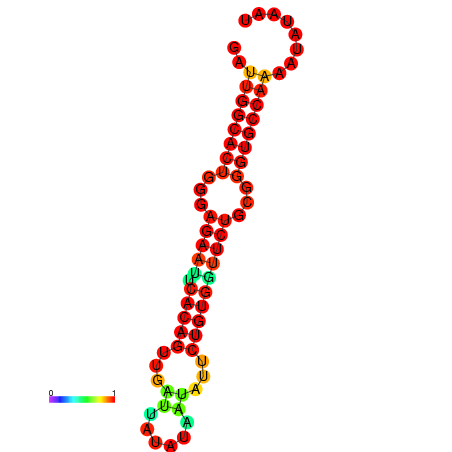 |
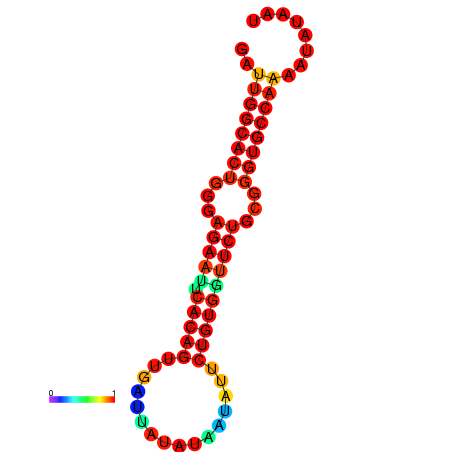 |
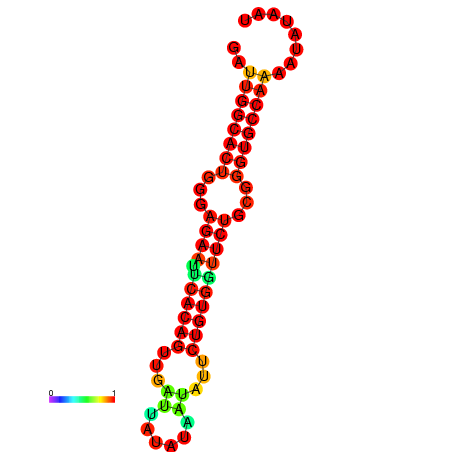 |
| dG=-26.8, p-value=0.009901 | dG=-26.8, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-26.0, p-value=0.009901 |
|---|---|---|---|
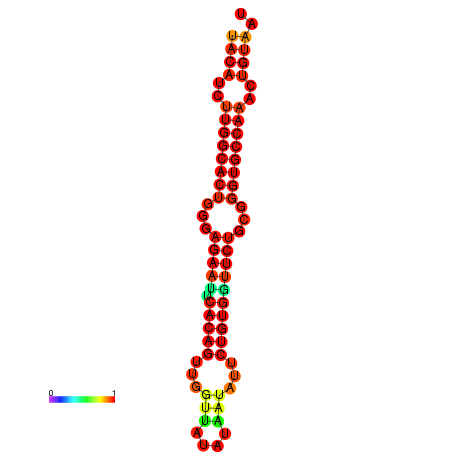 |
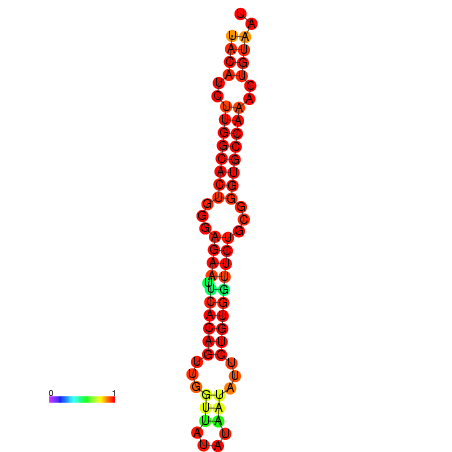 |
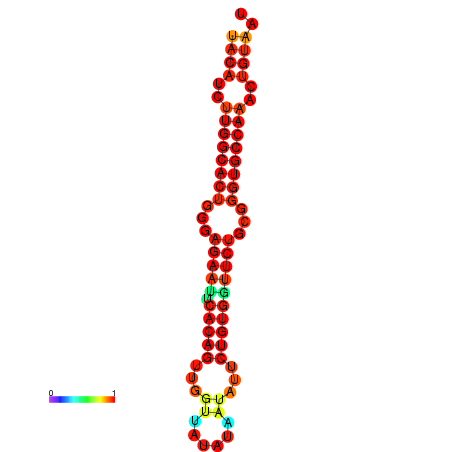 |
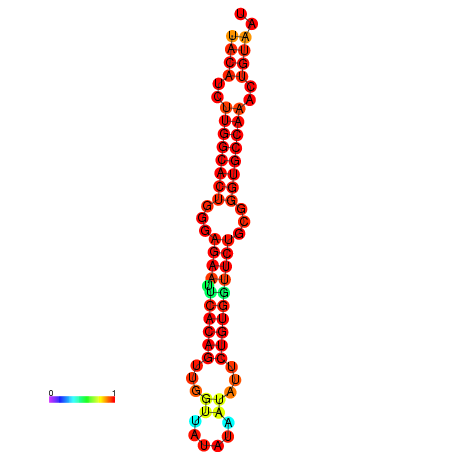 |
Generated: 03/07/2013 at 05:01 PM