| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:11953395-11953518 - | CCATCTAAT----------------------------------------GGT--ATATAGATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTAATTTTTA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGAGAATAAAACAT---------CCAAAATGAA |
| droSim1 | chr2L:11760450-11760573 - | CCATCTAAT----------------------------------------GGT--ATATAGATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTAATTTTTA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGAGAATAAAACAT---------CCAAAATGAA |
| droSec1 | super_16:152684-152807 - | CCTTCTAAT----------------------------------------GGT--ATATAGATCTCGACACAGTTAATGGCACTGGAAGAATTCACGGGGTAATTTTTA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGAGAATAAAACAT---------CCAAAGTGAA |
| droYak2 | chr2L:8374818-8374940 - | CCAGCTGCT----------------------------------------GGT--ATATAGATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTACTTTTTA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGAGAATAAAACAT---------CCAA-ATGAA |
| droEre2 | scaffold_4929:13456433-13456555 + | CCATCTGAT----------------------------------------GGT--GTATAGATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTACTTTTGA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGAGAATAAAACAT---------CCGA-ATGAA |
| droAna3 | scaffold_12943:2289700-2289819 + | ACA----------------------------------------------AGT--TTCTCCATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTAATTTCTA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGGGATTAAAACATCAAA------A-AAGTGA- |
| dp4 | chr4_group3:7322788-7322905 + | ACTCATAAG----------------------------------------TGT--ATCCACATCCCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTT-TTTCAA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGGGACT----CAT---------CTCA-ATCAA |
| droPer1 | super_1:8768281-8768398 + | ACTCATAAG----------------------------------------TGT--ATCCACATCCCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTT-TTTCAA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGGGACT----CAT---------CTCA-ATCAA |
| droWil1 | scaffold_180708:2657721-2657853 - | CTTTCGATT----------------------------------------GG---TTCTGCATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTTTTTATTTAACAC-ACCCGTGATCTCTTAGTGGCATCTATGGTGCGGGATTTAAGCATACAGAACATCCGACATCAA |
| droVir3 | scaffold_12963:13235091-13235216 + | CTTAATGTT----------------------------------------GCTTGCTCTGCATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTTTTTATAA--CAC-GCCCGTGATCTCTTAGTGGCATCTATGGTGCGGGA-TATTACGTACAA------C-AAATGAA |
| droMoj3 | Unknown | ACT-------------------------------------------------------------------AGTGATTGGCACTGGGAGAATTCACAGTTGA-TTATAT--AATATTCTGTGGTTCTGC-----------GGGTGCCAAAATATAATAT---------TCGGAATAAT |
| droGri2 | scaffold_15126:6621358-6621517 - | CTACCTGAATTTCTGCTGGGTAAGTTATTTTTTTCAAGTGATTTTTCTTGTT-GTTCTGCATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTTATTTTAA--CAC-GCCCGTGATCTCTTAGTGGCATCTATGGTGCGGGA-TATTACGT---------GCAGA---AA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:11953395-11953518 - | CCATCTAAT----------------------------------------GGT--ATATAGATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTAATTTTTA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGAGAATAAAACAT---------CCAAAATGAA |
| droSim1 | chr2L:11760450-11760573 - | CCATCTAAT----------------------------------------GGT--ATATAGATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTAATTTTTA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGAGAATAAAACAT---------CCAAAATGAA |
| droSec1 | super_16:152684-152807 - | CCTTCTAAT----------------------------------------GGT--ATATAGATCTCGACACAGTTAATGGCACTGGAAGAATTCACGGGGTAATTTTTA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGAGAATAAAACAT---------CCAAAGTGAA |
| droYak2 | chr2L:8374818-8374940 - | CCAGCTGCT----------------------------------------GGT--ATATAGATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTACTTTTTA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGAGAATAAAACAT---------CCAA-ATGAA |
| droEre2 | scaffold_4929:13456433-13456555 + | CCATCTGAT----------------------------------------GGT--GTATAGATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTACTTTTGA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGAGAATAAAACAT---------CCGA-ATGAA |
| droAna3 | scaffold_12943:2289700-2289819 + | ACA----------------------------------------------AGT--TTCTCCATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTAATTTCTA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGGGATTAAAACATCAAA------A-AAGTGA- |
| dp4 | chr4_group3:7322788-7322905 + | ACTCATAAG----------------------------------------TGT--ATCCACATCCCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTT-TTTCAA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGGGACT----CAT---------CTCA-ATCAA |
| droPer1 | super_1:8768281-8768398 + | ACTCATAAG----------------------------------------TGT--ATCCACATCCCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTT-TTTCAA--TACAACCCGTGATCTCTTAGTGGCATCTATGGTGCGGGACT----CAT---------CTCA-ATCAA |
| droWil1 | scaffold_180708:2657721-2657853 - | CTTTCGATT----------------------------------------GG---TTCTGCATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTTTTTATTTAACAC-ACCCGTGATCTCTTAGTGGCATCTATGGTGCGGGATTTAAGCATACAGAACATCCGACATCAA |
| droVir3 | scaffold_12963:13235091-13235216 + | CTTAATGTT----------------------------------------GCTTGCTCTGCATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTTTTTATAA--CAC-GCCCGTGATCTCTTAGTGGCATCTATGGTGCGGGA-TATTACGTACAA------C-AAATGAA |
| droMoj3 | Unknown | ACT-------------------------------------------------------------------AGTGATTGGCACTGGGAGAATTCACAGTTGA-TTATAT--AATATTCTGTGGTTCTGC-----------GGGTGCCAAAATATAATAT---------TCGGAATAAT |
| droGri2 | scaffold_15126:6621358-6621517 - | CTACCTGAATTTCTGCTGGGTAAGTTATTTTTTTCAAGTGATTTTTCTTGTT-GTTCTGCATCTCGGCACAGTTAATGGCACTGGAAGAATTCACGGGGTTATTTTAA--CAC-GCCCGTGATCTCTTAGTGGCATCTATGGTGCGGGA-TATTACGT---------GCAGA---AA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
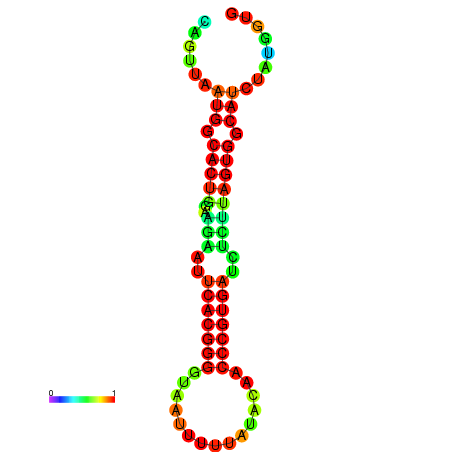 |
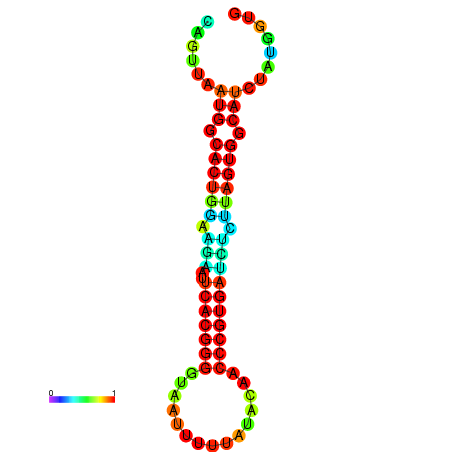
| dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
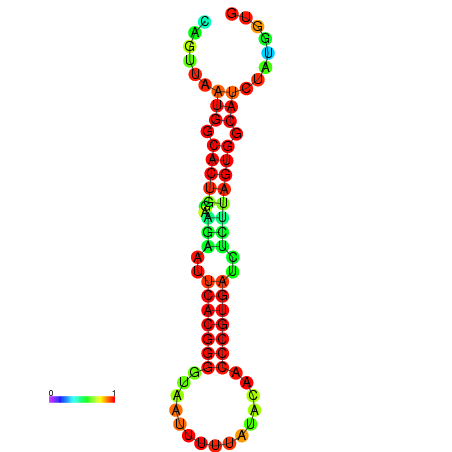 |
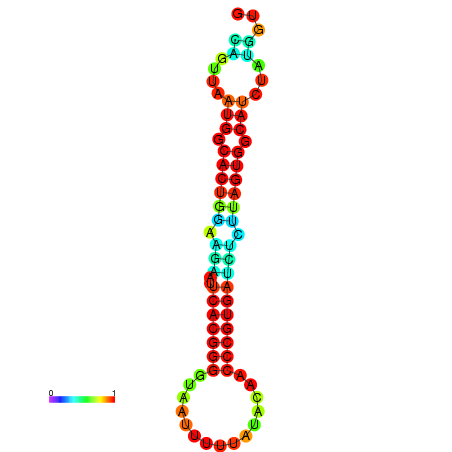
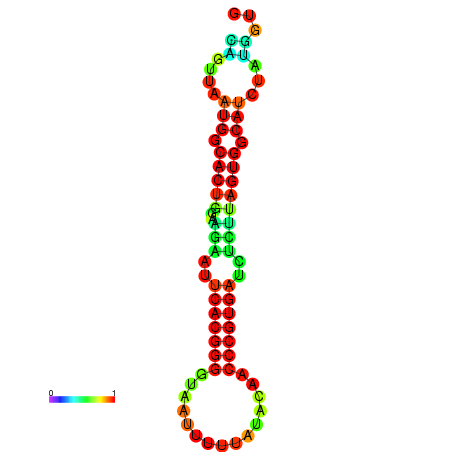
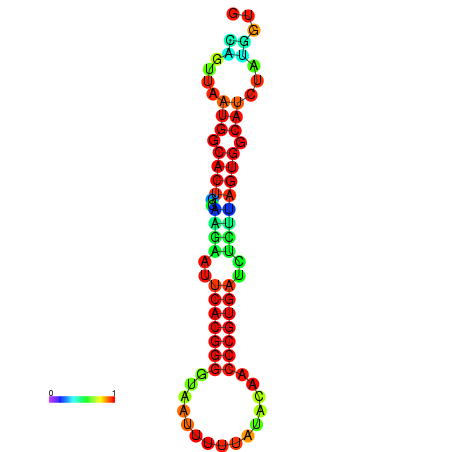
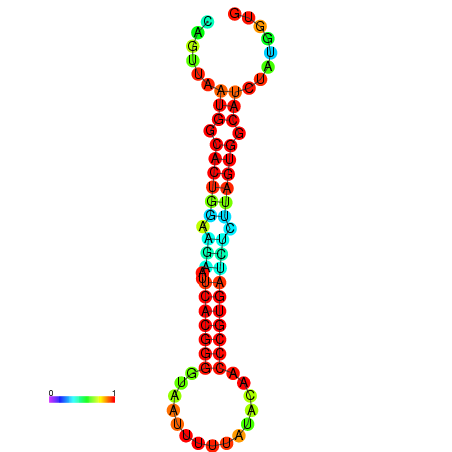
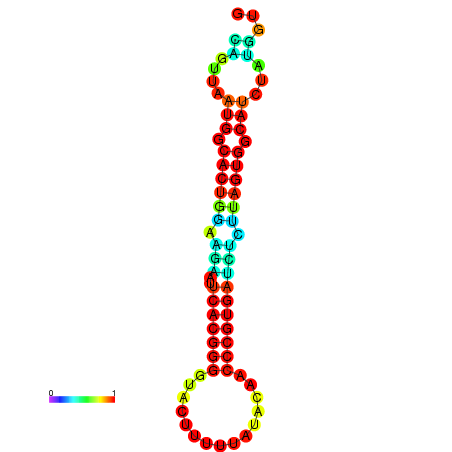
| dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
 |
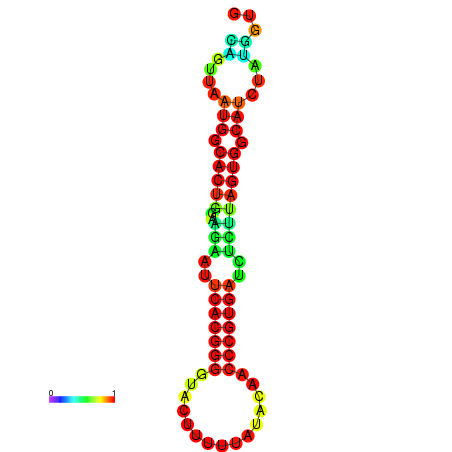 |
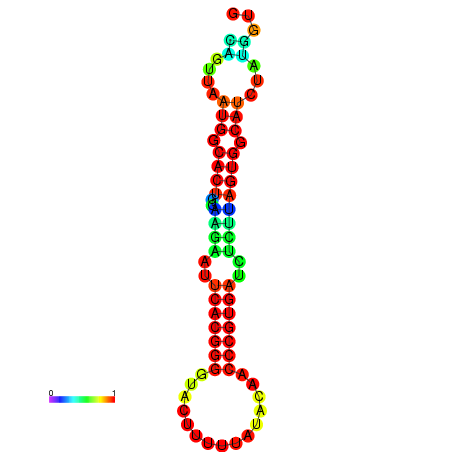 |
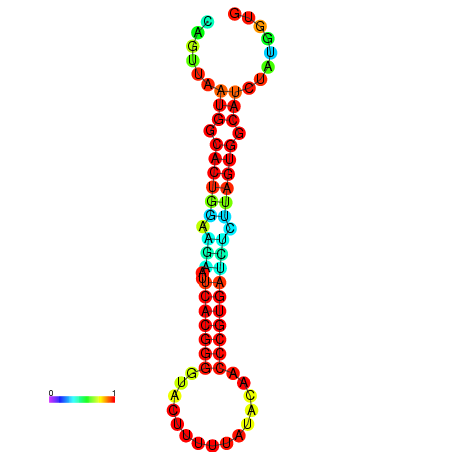 |
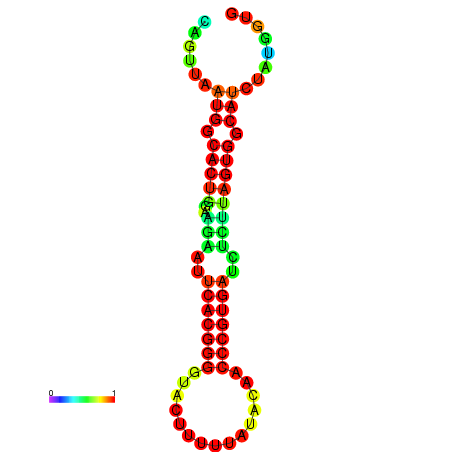 |
| dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
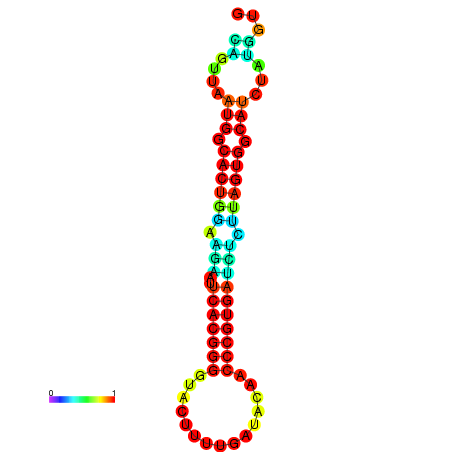 |
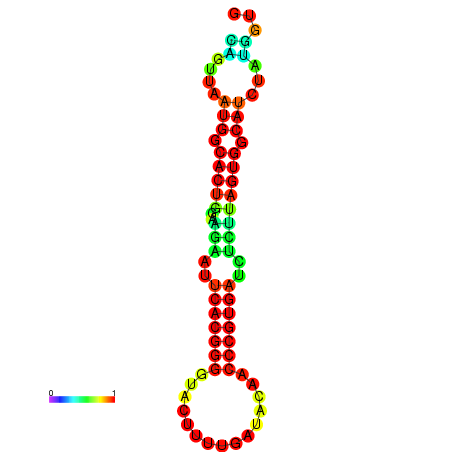 |
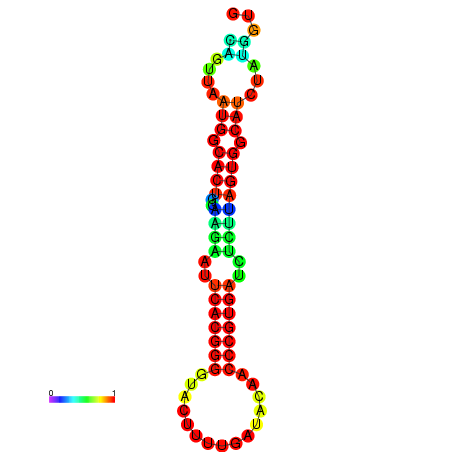 |
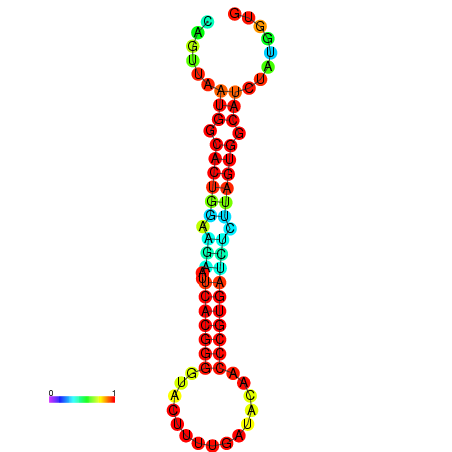 |
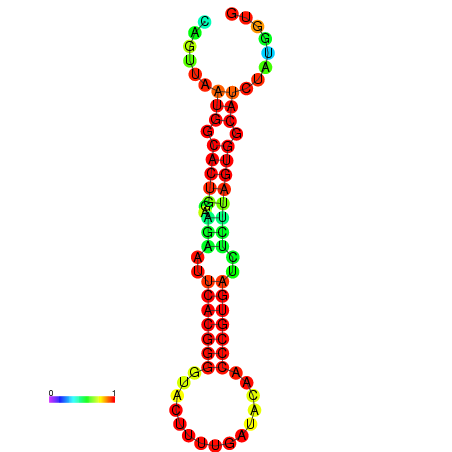 |
| dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
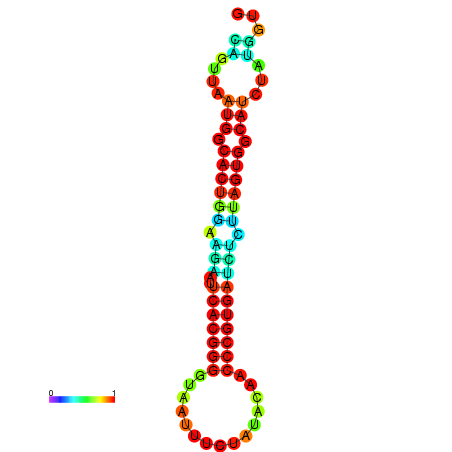 |
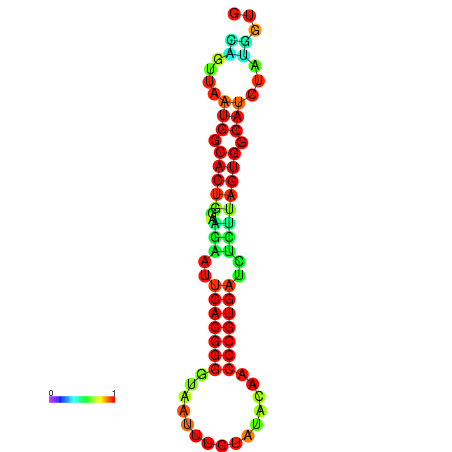 |
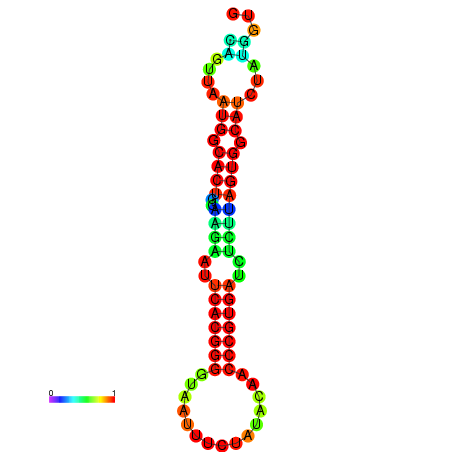 |
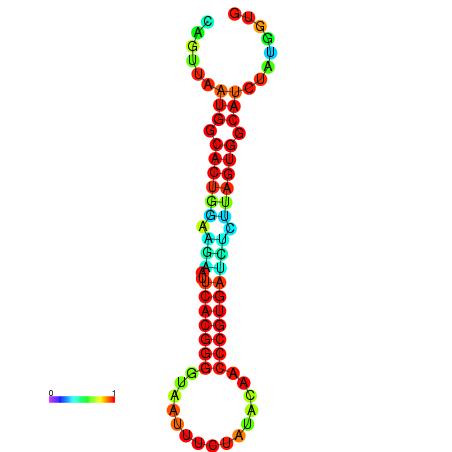 |
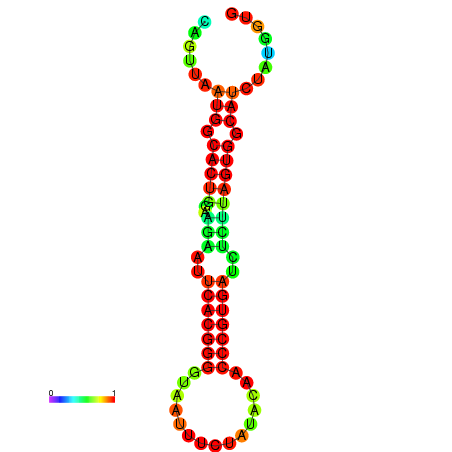 |
| dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
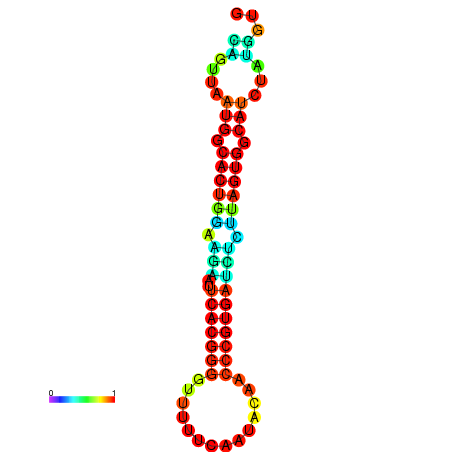 |
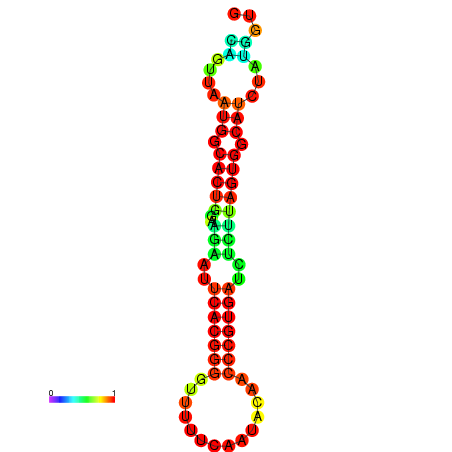 |
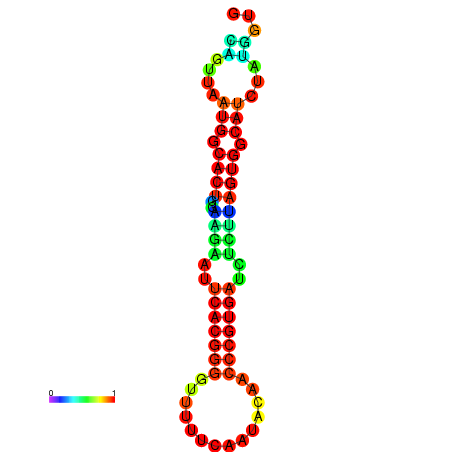 |
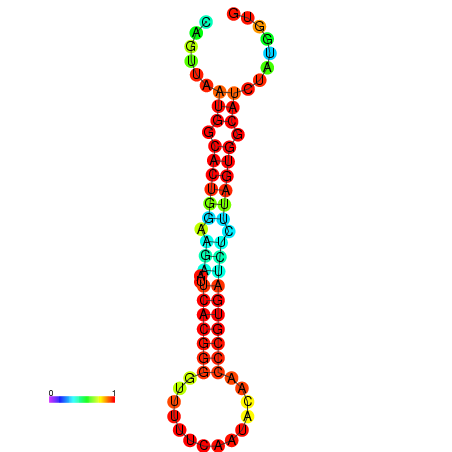 |
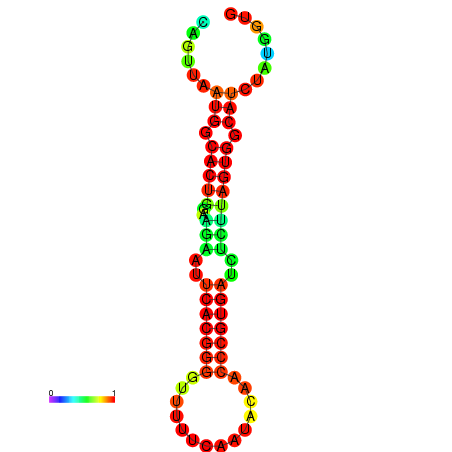 |
| dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
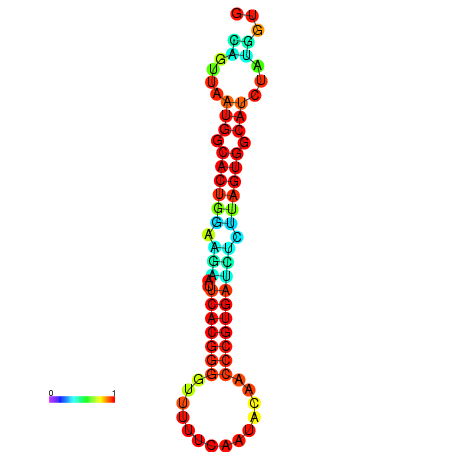 |
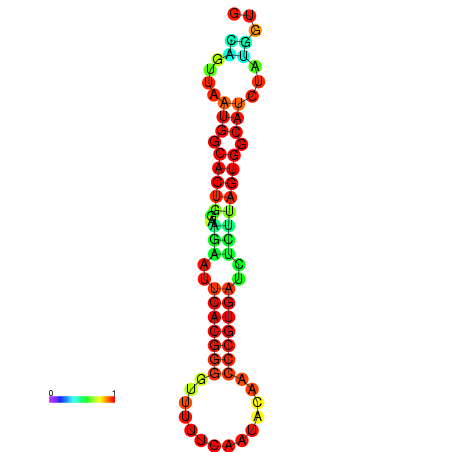 |
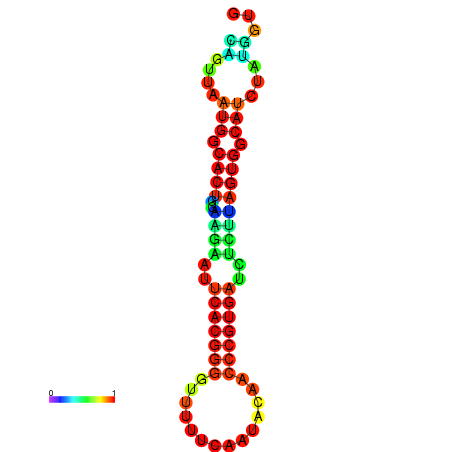 |
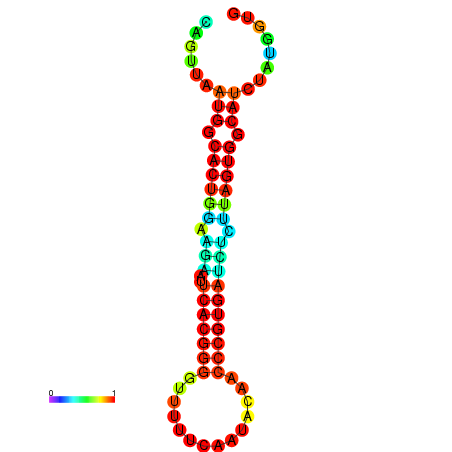 |
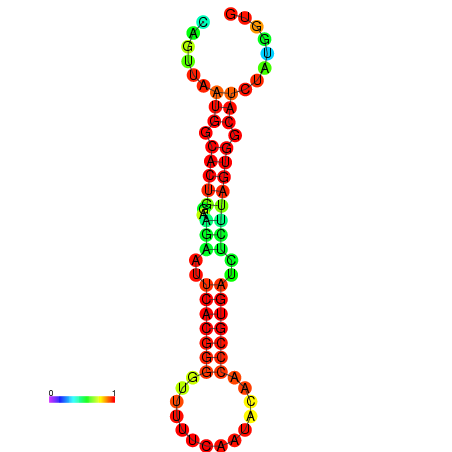 |
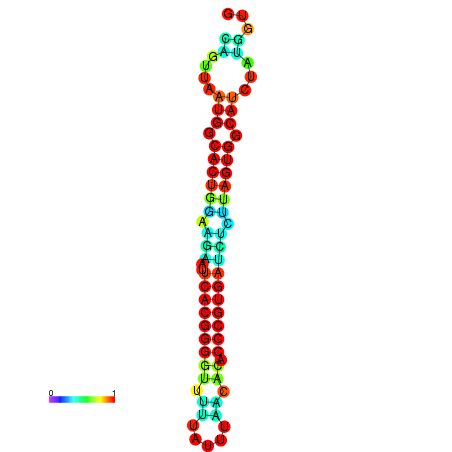
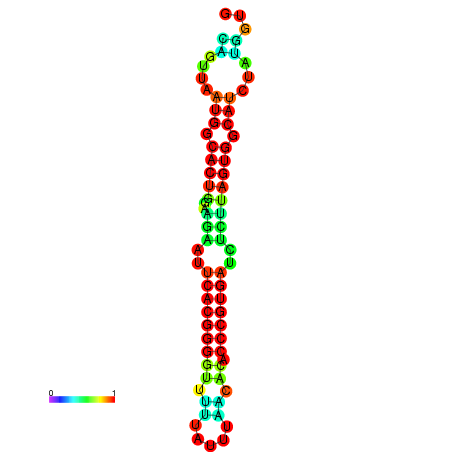
| dG=-24.5, p-value=0.009901 | dG=-24.4, p-value=0.009901 | dG=-24.3, p-value=0.009901 | dG=-24.3, p-value=0.009901 | dG=-24.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
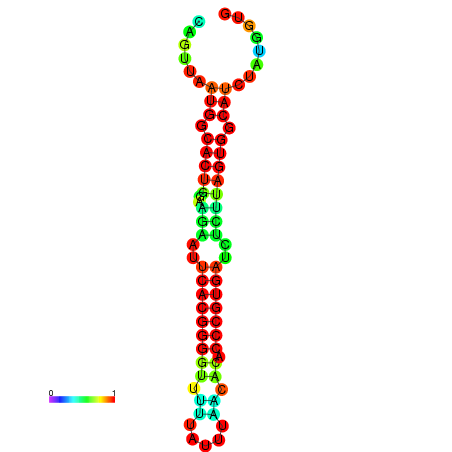 |
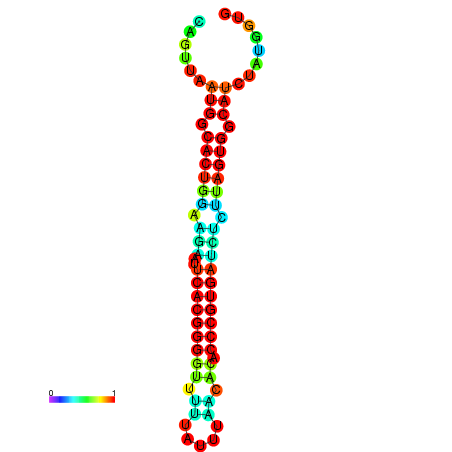
| dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.8, p-value=0.009901 |
|---|---|---|---|---|
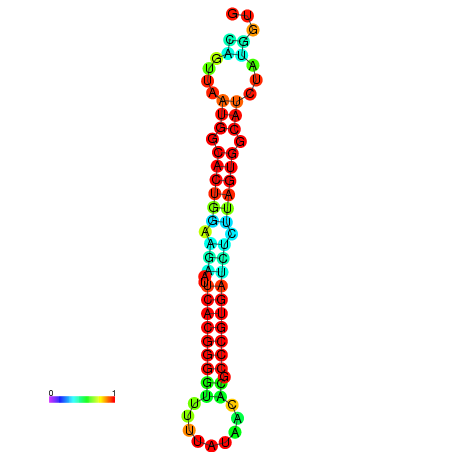 |
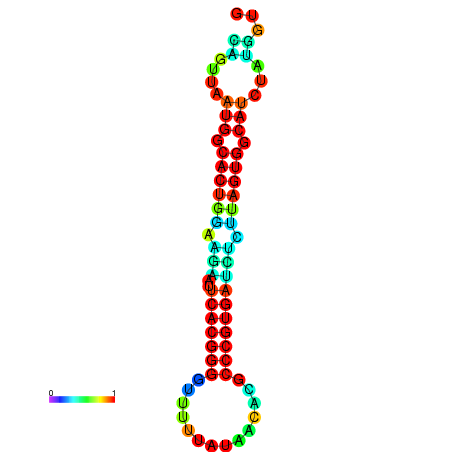 |
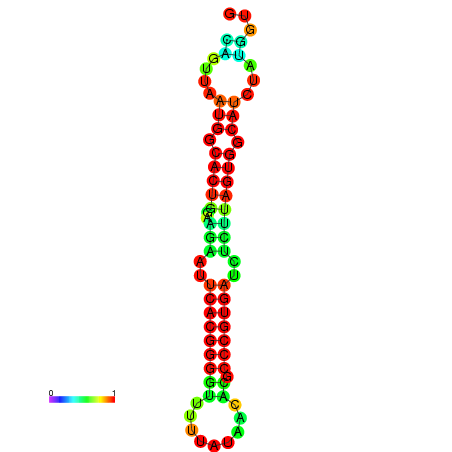 |
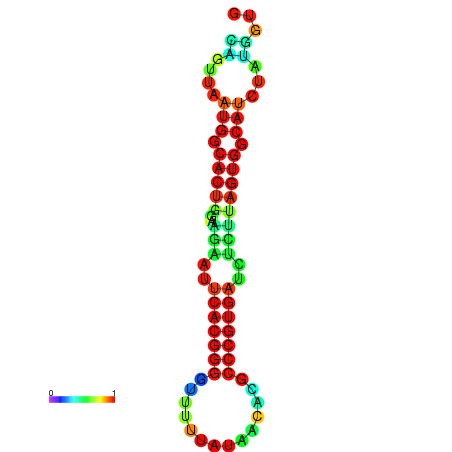 |
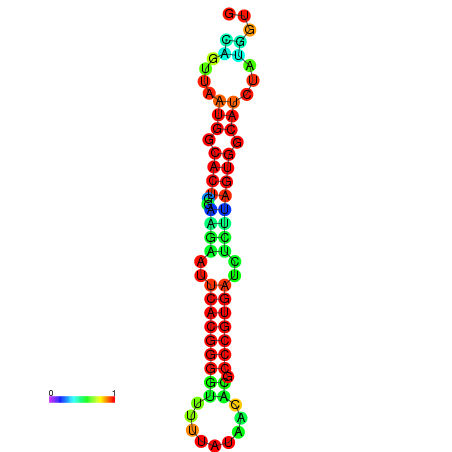 |
| dG=-13.4, p-value=0.009901 | dG=-13.4, p-value=0.009901 | dG=-12.6, p-value=0.009901 | dG=-12.6, p-value=0.009901 | dG=-12.6, p-value=0.009901 |
|---|---|---|---|---|
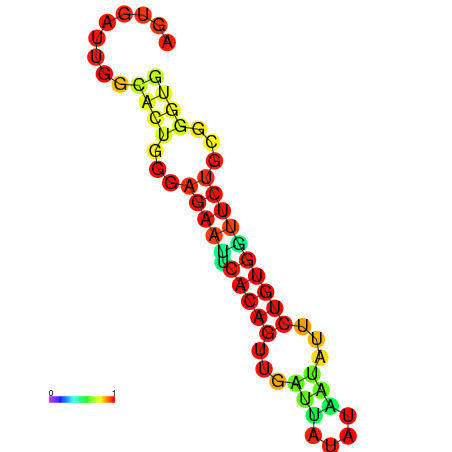 |
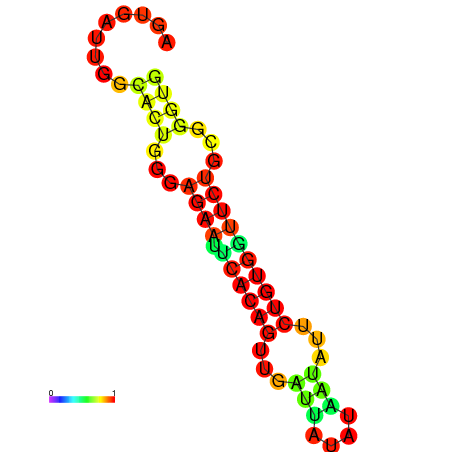 |
 |
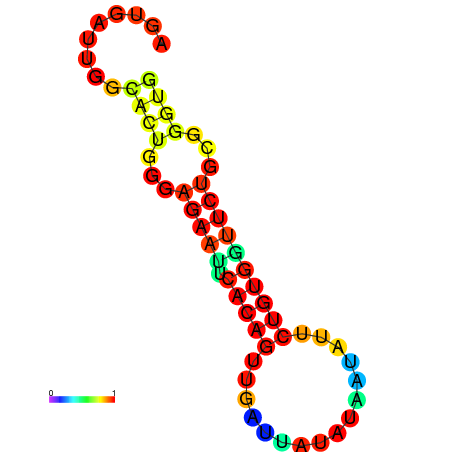 |
 |
| dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 05:01 PM