| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:21169287-21169387 - | ACGTACATCACTAAGGTAAGTGACTATGCCG-----GGATTGTGGTTAT---TGAACTTAGCG----TCTCCACT--CACGGC-ATTT--CAT------------TCACTTGCAGTACATCACCGCCGAG |
| droSim1 | chrX:12918307-12918407 + | ACGTACATCACCAAGGTAAGTGACTATGCCG-----GGATTGTGGTTAT---TATACTTAGCG----TCTCCACT--CACGGC-ATTT--CAT------------TCACTTGCAGTACATCACCGCCGAG |
| droSec1 | super_8:3476294-3476394 - | ACGTACATCACCAAGGTAAGTGACTATGCCG-----GGATTGTGGTTAT---TATACTTAGAG----TCTCCACT--CACGGC-ATTT--CAT------------TCACTTGCAGTACATCACCGCCGAG |
| droYak2 | chrX:20345010-20345110 - | ACGTACATCACCAAGGTAAGTGACAATGCCG-----GGATTGTGGTTAT---TGAAAATACCT----TCTCCACT--CACGGC-ATTA--TAT------------TCACTTGCAGTACATCACCGCCGAG |
| droEre2 | scaffold_4690:17698644-17698744 - | ACGTACATCACCAAGGTAAGTGACAGTGCCG-----GGATTGTGGTTAT---TAAAAATACCT----TCTCCACT--CACGGC-ATTT--CAT------------TCACTTGCAGTACATCACCGCCGAG |
| droAna3 | scaffold_13334:1053972-1054064 + | TCTGGCTACCGATTGGTGAGTTATCG----A-----CATTATCGATTTCTATCGATTT----------------------------CTATCGAGTTAAAAAAATCCCCTTTCCAGATCTGTGTCAAACGC |
| dp4 | chrXL_group1a:9058690-9058790 + | ACGTACATCACAAAGGTAAGTGGCTATGCCGCGACGGGATTG------------AACCTCTCATCTCTCACCTCT--CACGGC-ATAT--CAA------------TCACTTGCAGTACATCACCGCCGAG |
| droPer1 | super_15:1452144-1452244 + | ACGTACATCACAAAGGTAAGTGGCTATGCCGCGACGGGATTG------------AACCTCTCATCTCTCACCTCT--CACGGC-ATAT--CAA------------TCACTTGCAGTACATCACCGCCGAG |
| droWil1 | Unknown | ACTTACATTACCAAGGTGAGT----------------------------------------------------------------TTT--GTT------------TTTCTTACAGTACATTACATCGGAG |
| droVir3 | Unknown | ACGTGCATTACCAAGGTAAGC-----------------------------------------------CTCTTCT--CACGAC-ATCCCTTAA------------TCCCTTGCAGTACGTTGCCAACGAA |
| droMoj3 | Unknown | ACATGCATAACCAGGGTAAGTGCTCA-GCTG-----CTtTTtTttTttT---tgttTTTTttT-------------------T-tTTT--CGT------------CGCCTTACAGTACATCTCCAAGGAG |
| droGri2 | scaffold_15203:11132616-11132716 + | ACCTACATTACTAAGGTATGT------TCGA-----TGATTTTGGTTTTTTTTGGCTTTACTT----TCACCGTTTACACGACAATTT--CAT------------CCTTTTGCAGTACGTGACCAGTGAG |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:21169287-21169387 - | ACGTACATCACTAAGGTAAGTGACTATGCCG-----GGATTGTGGTTATTGAACTTAGCG----TCTCCACTCACGGCATTTCATTCACTTGCAGTACATCACCGCCGAG |
| droSim1 | chrX:12918307-12918407 + | ACGTACATCACCAAGGTAAGTGACTATGCCG-----GGATTGTGGTTATTATACTTAGCG----TCTCCACTCACGGCATTTCATTCACTTGCAGTACATCACCGCCGAG |
| droSec1 | super_8:3476294-3476394 - | ACGTACATCACCAAGGTAAGTGACTATGCCG-----GGATTGTGGTTATTATACTTAGAG----TCTCCACTCACGGCATTTCATTCACTTGCAGTACATCACCGCCGAG |
| droYak2 | chrX:20345010-20345110 - | ACGTACATCACCAAGGTAAGTGACAATGCCG-----GGATTGTGGTTATTGAAAATACCT----TCTCCACTCACGGCATTATATTCACTTGCAGTACATCACCGCCGAG |
| droEre2 | scaffold_4690:17698644-17698744 - | ACGTACATCACCAAGGTAAGTGACAGTGCCG-----GGATTGTGGTTATTAAAAATACCT----TCTCCACTCACGGCATTTCATTCACTTGCAGTACATCACCGCCGAG |
| dp4 | chrXL_group1a:9058690-9058790 + | ACGTACATCACAAAGGTAAGTGGCTATGCCGCGACGGGATTG---------AACCTCTCATCTCTCACCTCTCACGGCATATCAATCACTTGCAGTACATCACCGCCGAG |
| droPer1 | super_15:1452144-1452244 + | ACGTACATCACAAAGGTAAGTGGCTATGCCGCGACGGGATTG---------AACCTCTCATCTCTCACCTCTCACGGCATATCAATCACTTGCAGTACATCACCGCCGAG |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-17.7, p-value=0.009901 | dG=-17.7, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-17.7, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.4, p-value=0.009901 | dG=-17.4, p-value=0.009901 | dG=-17.3, p-value=0.009901 |
|---|---|---|---|---|
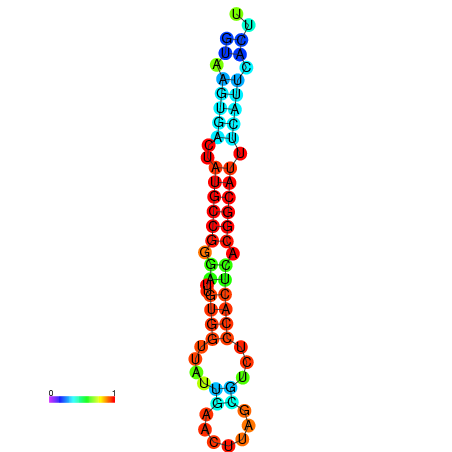
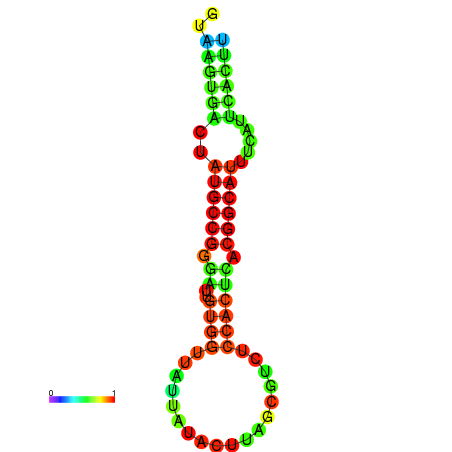 |
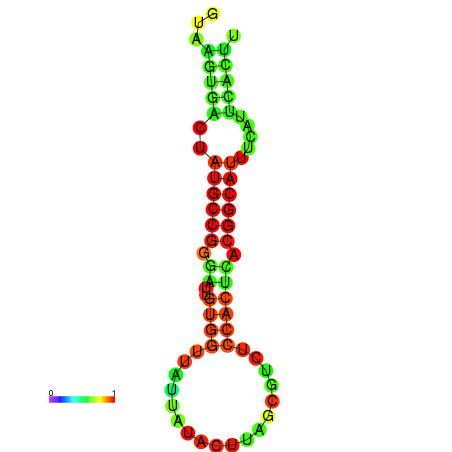 |
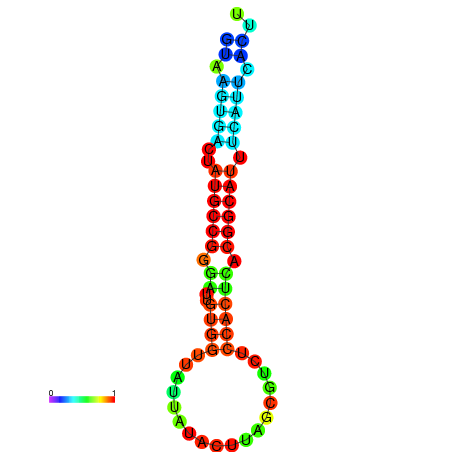 |
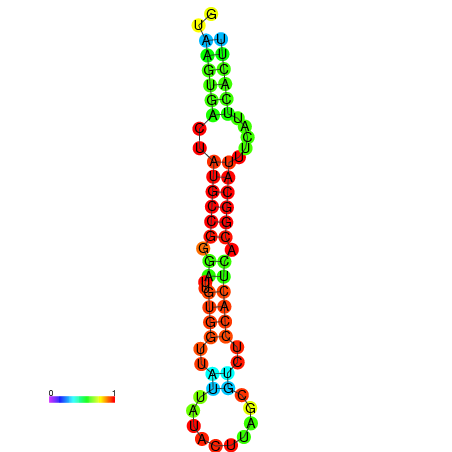 |
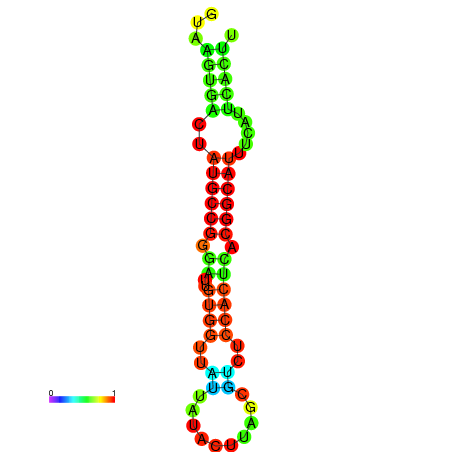 |
| dG=-19.4, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.1, p-value=0.009901 | dG=-18.9, p-value=0.009901 | dG=-18.8, p-value=0.009901 |
|---|---|---|---|---|
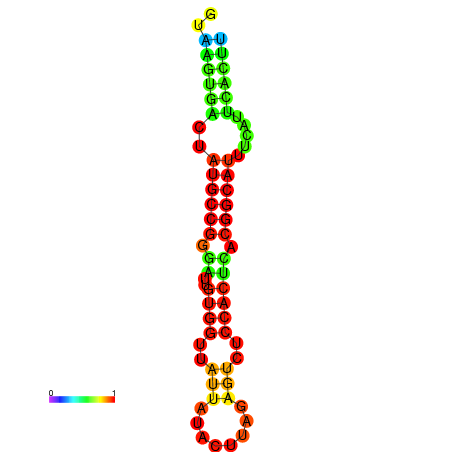 |
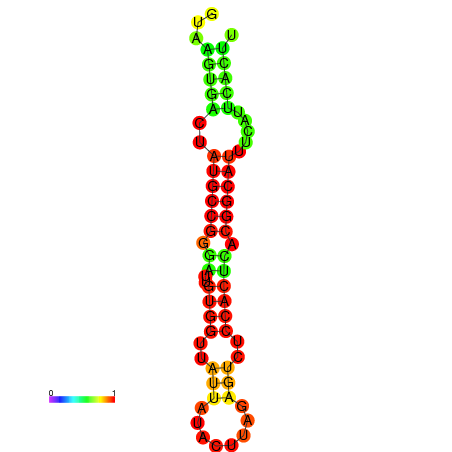 |
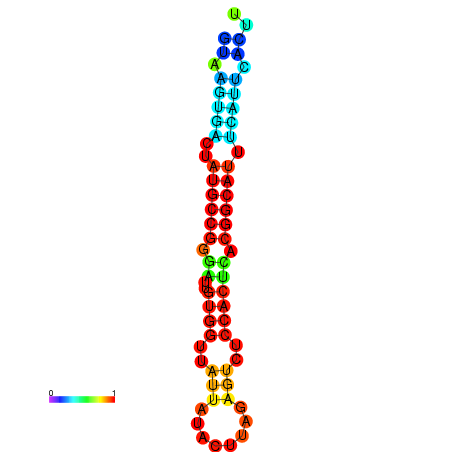 |
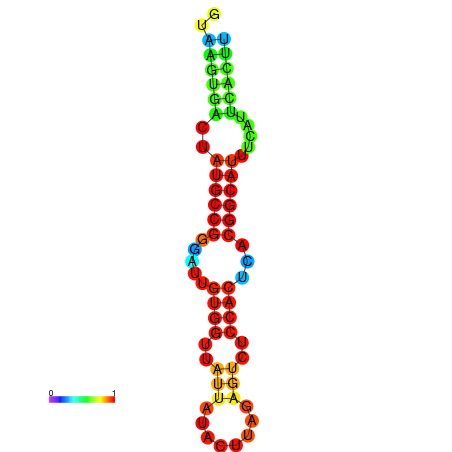 |
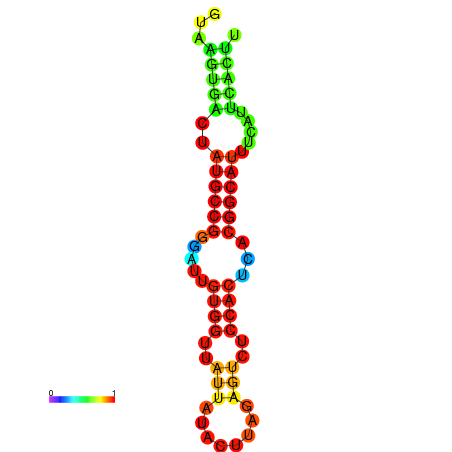 |
| dG=-18.1, p-value=0.009901 | dG=-18.0, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.5, p-value=0.009901 |
|---|---|---|---|---|
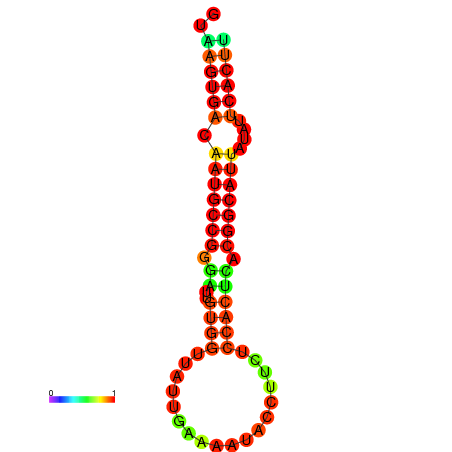 |
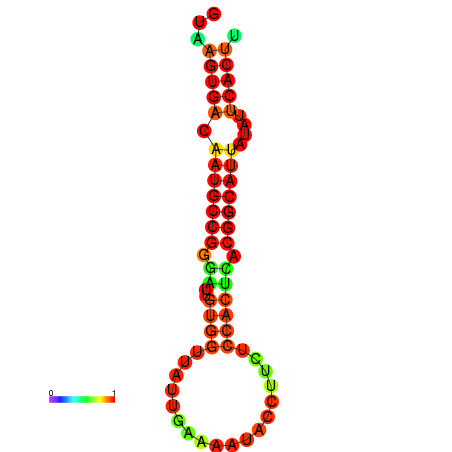 |
 |
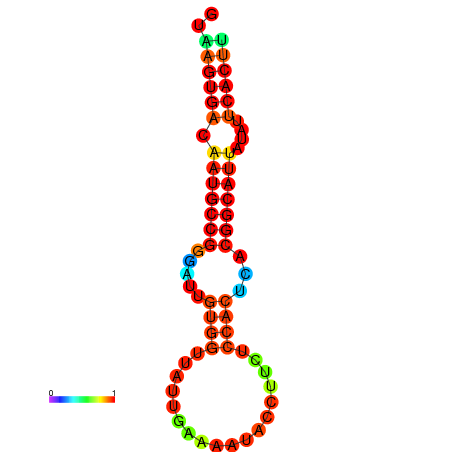 |
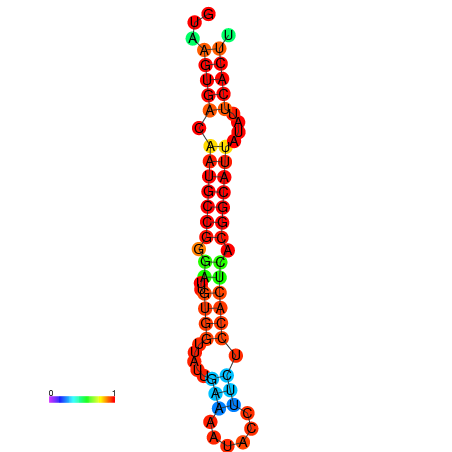 |
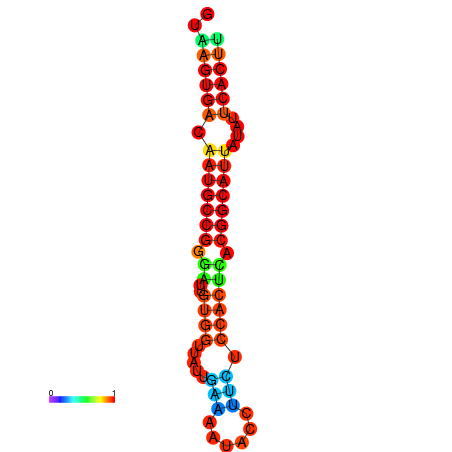
| dG=-18.4, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.0, p-value=0.009901 | dG=-18.0, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|---|---|---|
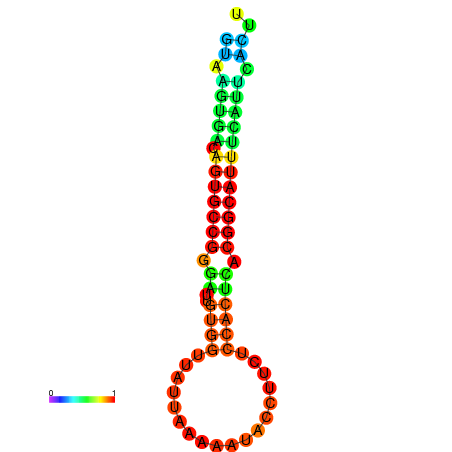 |
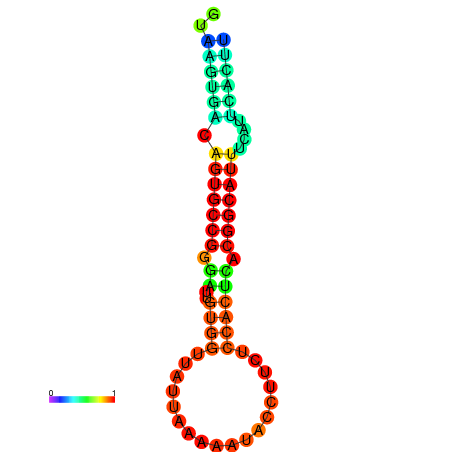 |
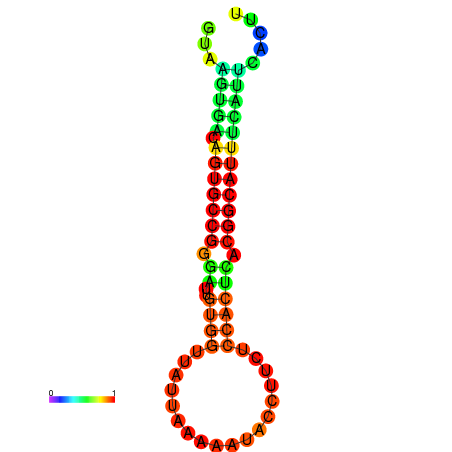 |
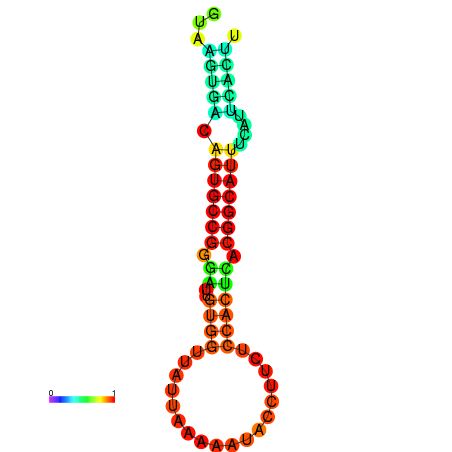 |
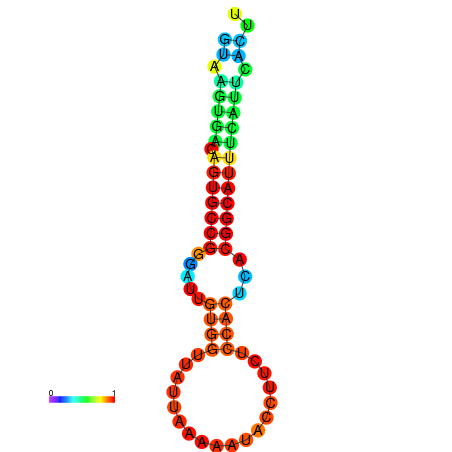 |
| dG=-9.0, p-value=0.009901 | dG=-8.7, p-value=0.009901 |
|---|---|
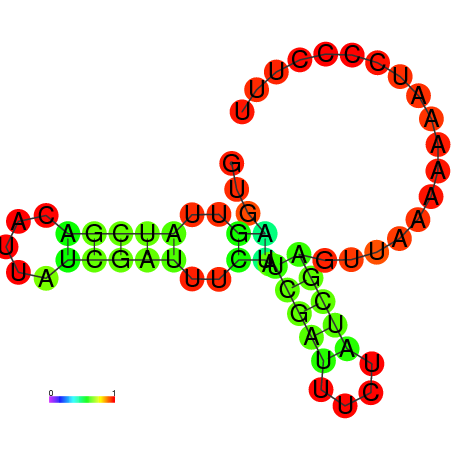 |
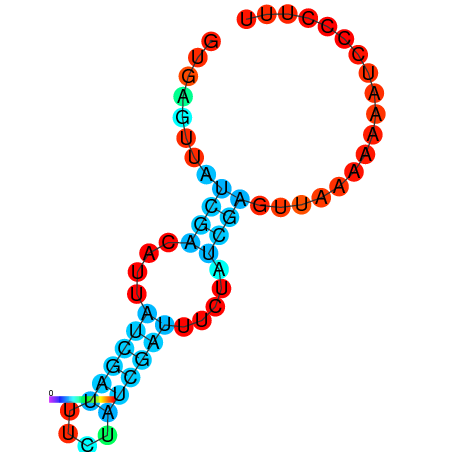 |
| dG=-18.2, p-value=0.009901 | dG=-18.2, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 |
|---|---|---|---|---|
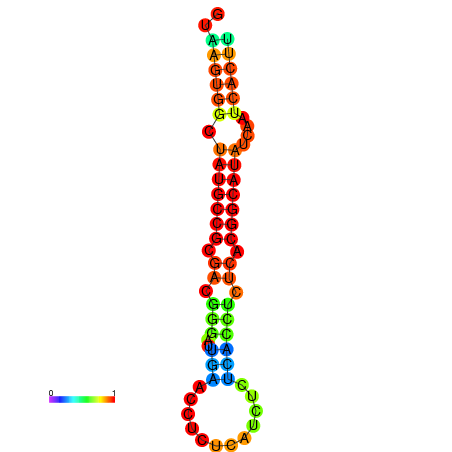 |
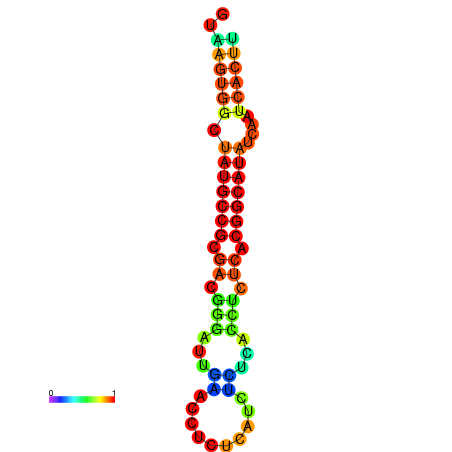 |
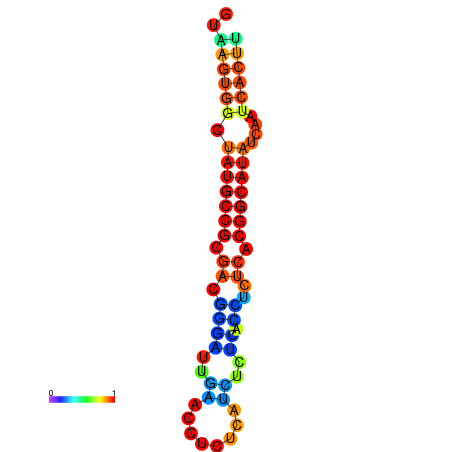 |
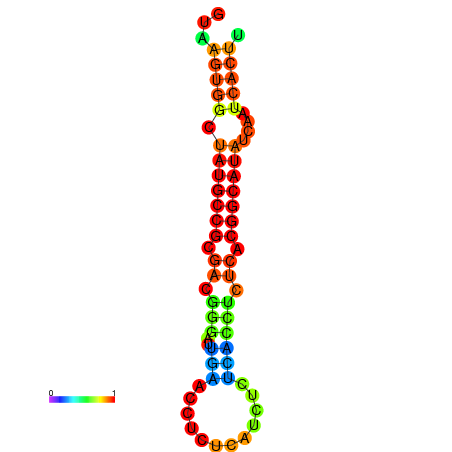 |
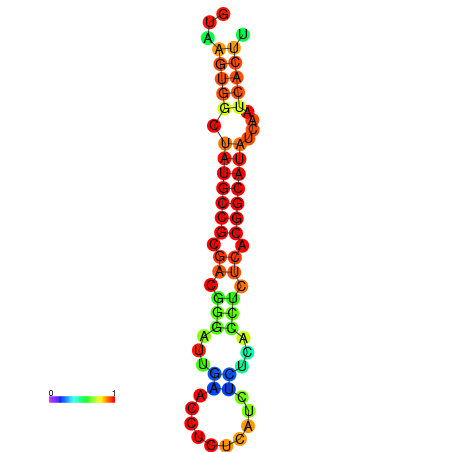 |
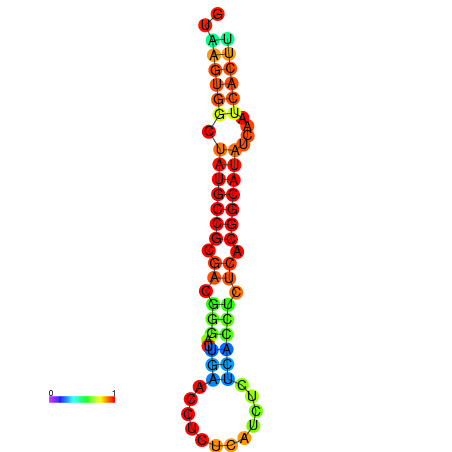
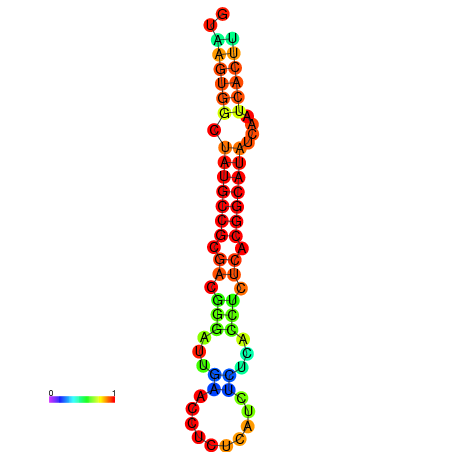
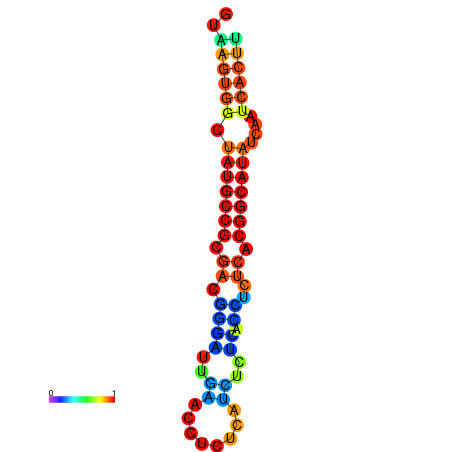
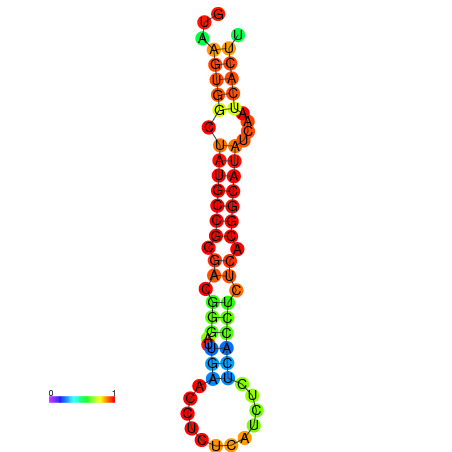
| dG=-18.2, p-value=0.009901 | dG=-18.2, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=0.0, p-value=1.0 | dG=0.8, p-value=1.0 |
|---|---|
 |
 |
| dG=0.0, p-value=1.0 | dG=0.1, p-value=1.0 |
|---|---|
 |
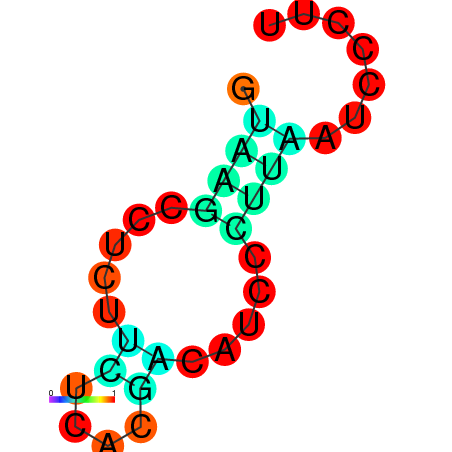 |
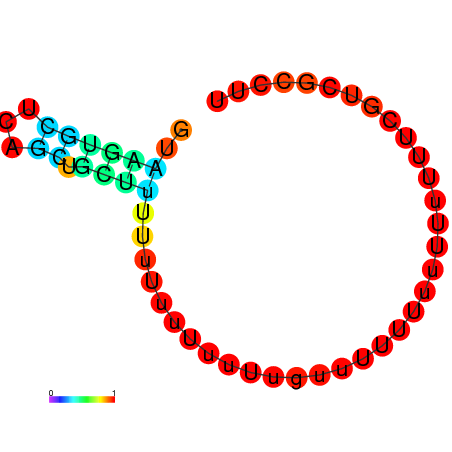
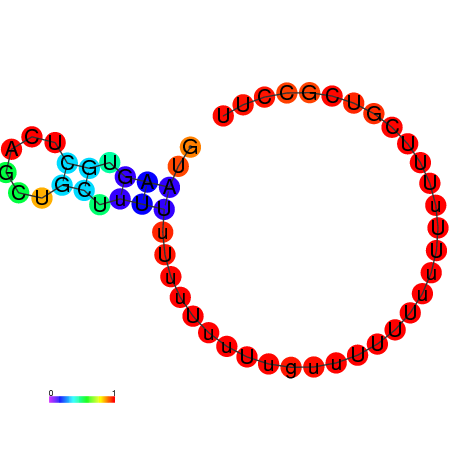
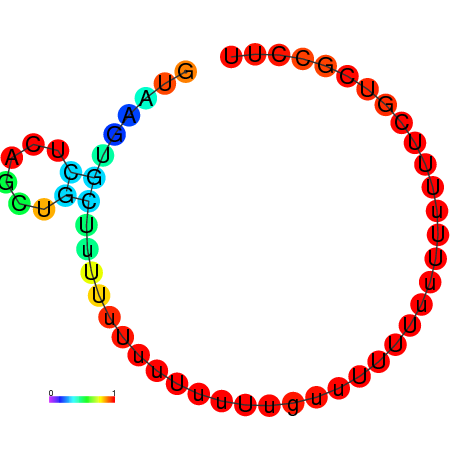
| dG=-1.4, p-value=0.019802 | dG=-0.8, p-value=0.019802 | dG=-0.8, p-value=0.009901 | dG=-0.7, p-value=0.009901 | dG=-0.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
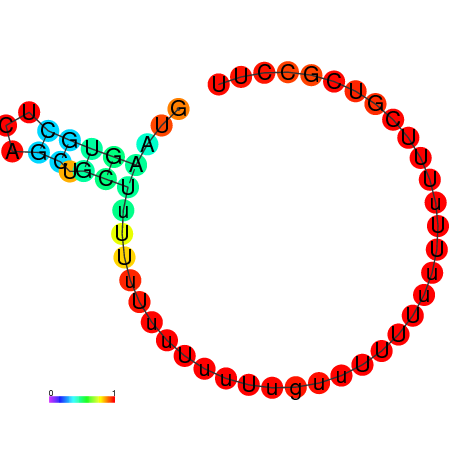 |
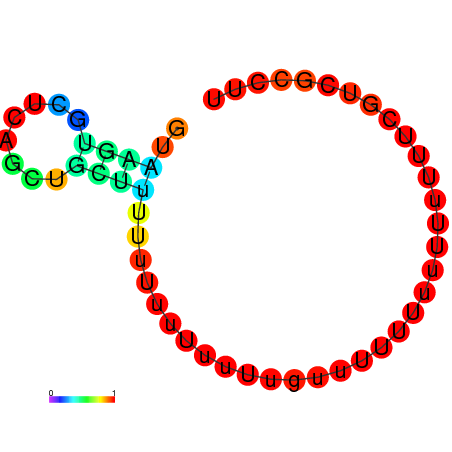 |
| dG=-7.5, p-value=0.009901 |
|---|
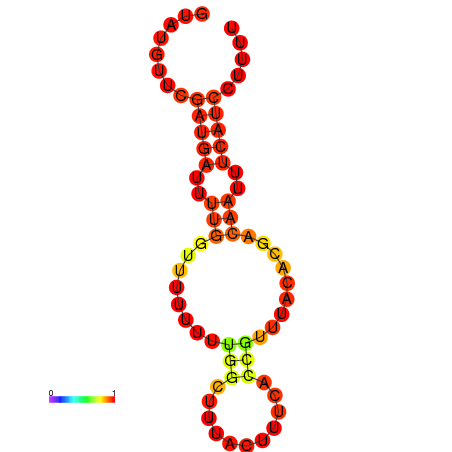 |
Generated: 03/07/2013 at 04:59 PM