
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:9289926-9290048 - | GTG-------TGCCG------------CCTACTTACC-----------------------------------------------AAGTTCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGCCCAGCAAAA---------GAA |
| droSim1 | chr3R_random:533427-533549 - | GTG-------TGCCG------------TCTACTTACC-----------------------------------------------AAGTTCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGCCCAGCAAAA---------GAA |
| droSec1 | super_0:12715202-12715324 + | GTG-------TGCCG------------TCTACTTACC-----------------------------------------------AAGTTCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGCCCAGCAAAA---------GAA |
| droYak2 | chr3R:13598006-13598135 - | GTGCTAGATGTACGA------------TGTACTTACC-----------------------------------------------AAGTTCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGCCCAGCAAAA---------GAA |
| droEre2 | scaffold_4770:12350666-12350788 + | GTG-------CTAGA------------TGTACTTACC-----------------------------------------------AAGTTCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGCCCAGCAAAA---------GAA |
| droAna3 | scaffold_13340:1848657-1848824 - | GAG--------------------------TACTTAGCCCCAACCATTGGTAAATATTGATTTTATACTTACATTACTTCCTGCGAAG-TCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGTCCAGCAAAATACAAG---GAA |
| dp4 | chr2:16736618-16736739 - | T-A-------TCTTG------------TGCATTTTGG-----------------------------------------------CGGTTTGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCG-GTGGTCCAACAGTG---------AAA |
| droPer1 | super_0:3659179-3659300 + | T-A-------TCTTG------------TGCATTTTGG-----------------------------------------------CGGTTTGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCG-GTGGTCCAACAGTG---------AAA |
| droWil1 | scaffold_181130:6363211-6363342 - | GAT-------GGATCAATACTTACTTGAAC------------------------------------------------GAAG--CGTATTGCTTTACTAAGTACTAGTGCCGCAGGAGTTTGGTTCGTGTCCGTAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGACGACTTTACCTACTTAA---------AAA |
| droVir3 | scaffold_12855:2861518-2861626 + | T----------------------------------------------------------------------------------------TGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTTGTGTCCGCAATGCCTCCTGCTGCCCAAGTGCTTATTAAAGCGACTAGTTTTCCACCAGCA-ACAATTAGAAA |
| droMoj3 | scaffold_6540:10349255-10349360 - | T----------------------------------------------------------------------------------------TGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGGTTGTGTCCGCAATGCCTCCTGCTGCCCAAGTGCTTATCAAAGCGACGAGTTTTTCAACAGCA-GCAAT---TAA |
| droGri2 | scaffold_14906:3807816-3807948 - | TTG-------TGGCA----CTTACTTGAGC----------------------------------------------TTGATG--CGTATTGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTTGTGTCCGCAATGCCTCCTGCTGCCCAAGTGCTTATTAAGGCGACGAATTTTCCAACAATGTGC------GAG |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:9289926-9290048 - | GTG-------TGCCG------------CCTACTTACC-----------------------------------------------AAGTTCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGCCCAGCAAAA---------GAA |
| droSim1 | chr3R_random:533427-533549 - | GTG-------TGCCG------------TCTACTTACC-----------------------------------------------AAGTTCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGCCCAGCAAAA---------GAA |
| droSec1 | super_0:12715202-12715324 + | GTG-------TGCCG------------TCTACTTACC-----------------------------------------------AAGTTCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGCCCAGCAAAA---------GAA |
| droYak2 | chr3R:13598006-13598135 - | GTGCTAGATGTACGA------------TGTACTTACC-----------------------------------------------AAGTTCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGCCCAGCAAAA---------GAA |
| droEre2 | scaffold_4770:12350666-12350788 + | GTG-------CTAGA------------TGTACTTACC-----------------------------------------------AAGTTCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGCCCAGCAAAA---------GAA |
| droAna3 | scaffold_13340:1848657-1848824 - | GAG--------------------------TACTTAGCCCCAACCATTGGTAAATATTGATTTTATACTTACATTACTTCCTGCGAAG-TCGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCGAGTTGTCCAGCAAAATACAAG---GAA |
| dp4 | chr2:16736618-16736739 - | T-A-------TCTTG------------TGCATTTTGG-----------------------------------------------CGGTTTGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCG-GTGGTCCAACAGTG---------AAA |
| droPer1 | super_0:3659179-3659300 + | T-A-------TCTTG------------TGCATTTTGG-----------------------------------------------CGGTTTGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTCGTGTCCGCAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGGCG-GTGGTCCAACAGTG---------AAA |
| droWil1 | scaffold_181130:6363211-6363342 - | GAT-------GGATCAATACTTACTTGAAC------------------------------------------------GAAG--CGTATTGCTTTACTAAGTACTAGTGCCGCAGGAGTTTGGTTCGTGTCCGTAATACCTCCTGCTGCCCAAGTGCTTATTAAAGCGACGACTTTACCTACTTAA---------AAA |
| droVir3 | scaffold_12855:2861518-2861626 + | T----------------------------------------------------------------------------------------TGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTTGTGTCCGCAATGCCTCCTGCTGCCCAAGTGCTTATTAAAGCGACTAGTTTTCCACCAGCA-ACAATTAGAAA |
| droMoj3 | scaffold_6540:10349255-10349360 - | T----------------------------------------------------------------------------------------TGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGGTTGTGTCCGCAATGCCTCCTGCTGCCCAAGTGCTTATCAAAGCGACGAGTTTTTCAACAGCA-GCAAT---TAA |
| droGri2 | scaffold_14906:3807816-3807948 - | TTG-------TGGCA----CTTACTTGAGC----------------------------------------------TTGATG--CGTATTGCTTTCCTAAGTACTAGTGCCGCAGGAGTTAGGTTTGTGTCCGCAATGCCTCCTGCTGCCCAAGTGCTTATTAAGGCGACGAATTTTCCAACAATGTGC------GAG |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-25.7, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-25.7, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 |
|---|---|---|---|---|
 |
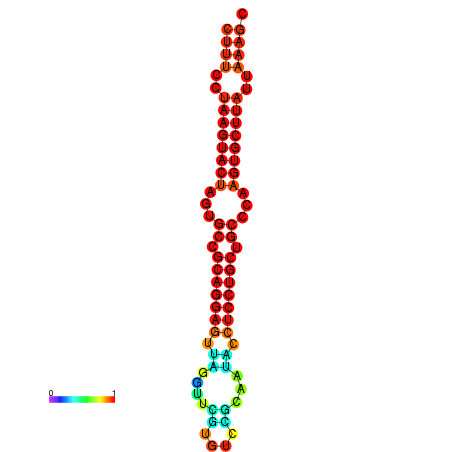 |
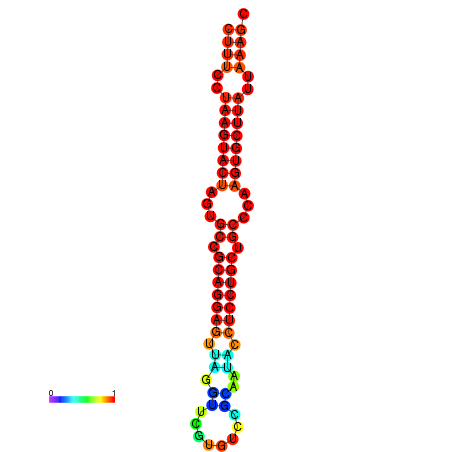 |
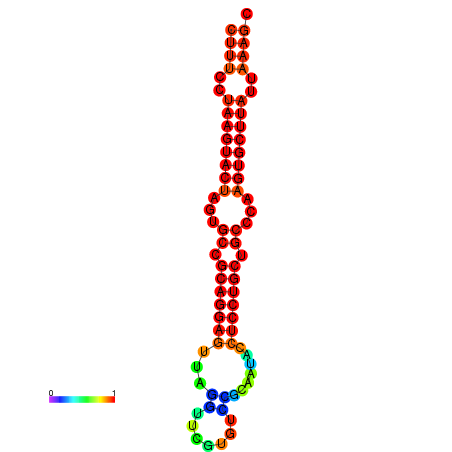 |
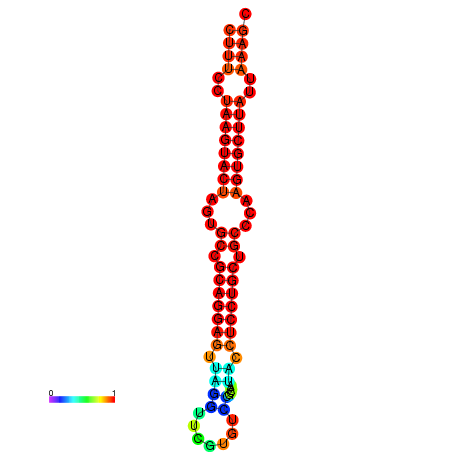 |
| dG=-25.7, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
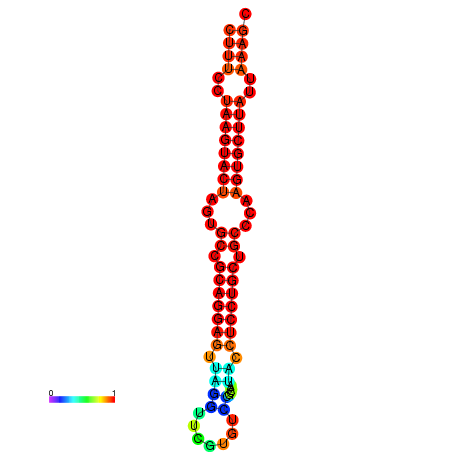 |
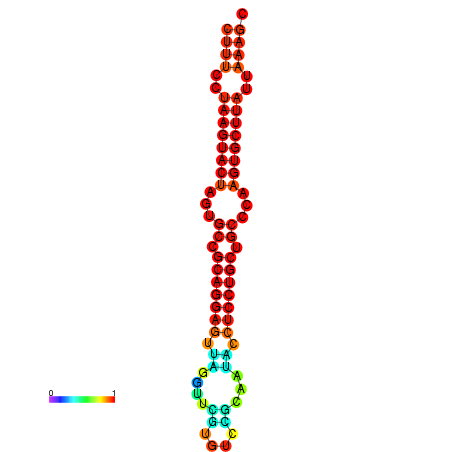
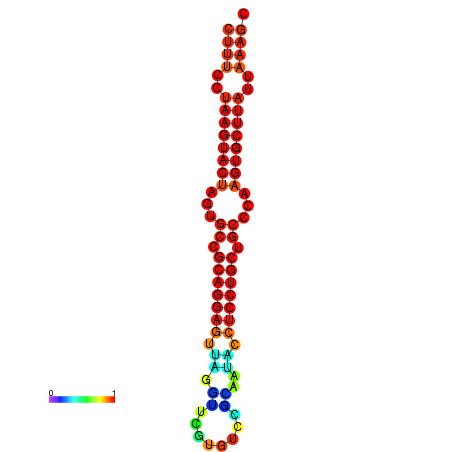
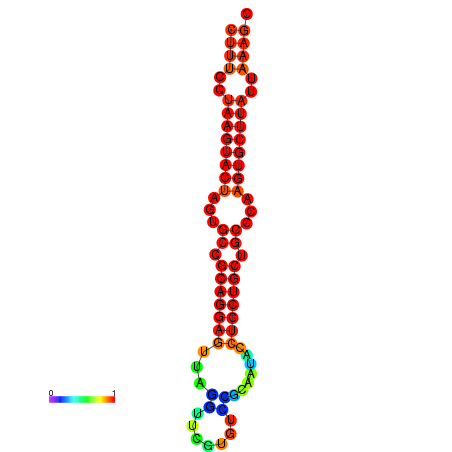
| dG=-25.7, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-25.7, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 |
|---|---|---|---|---|
 |
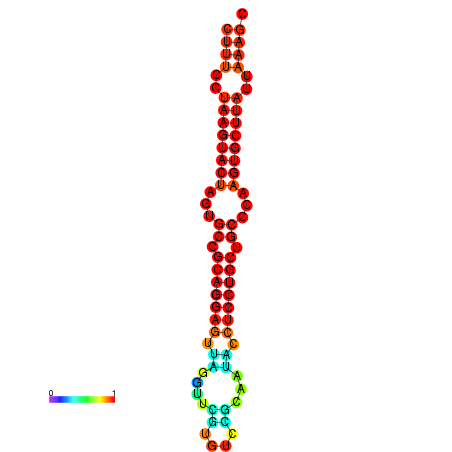 |
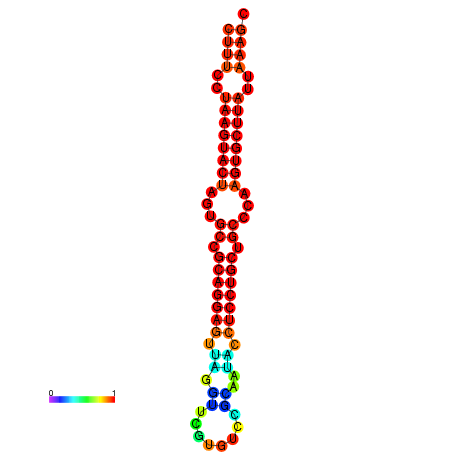 |
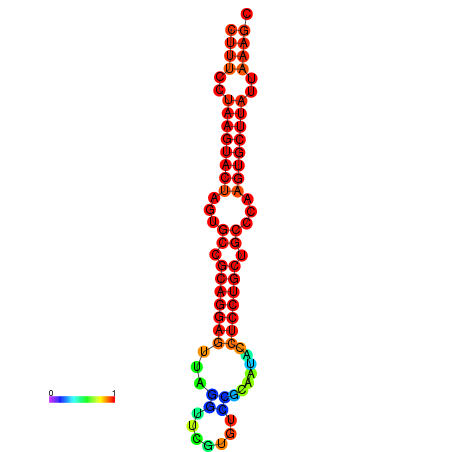 |
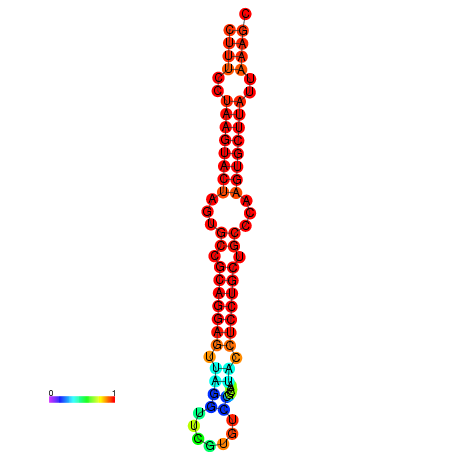 |
| dG=-25.7, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 |
|---|---|---|---|---|
 |
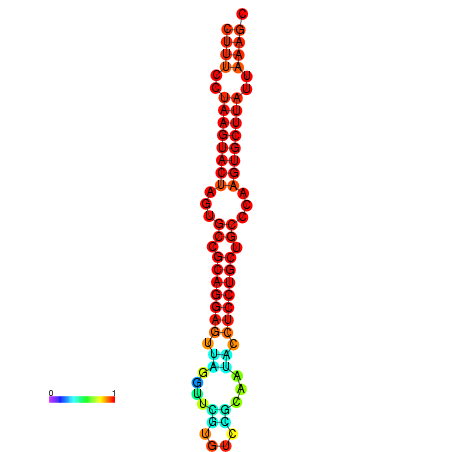 |
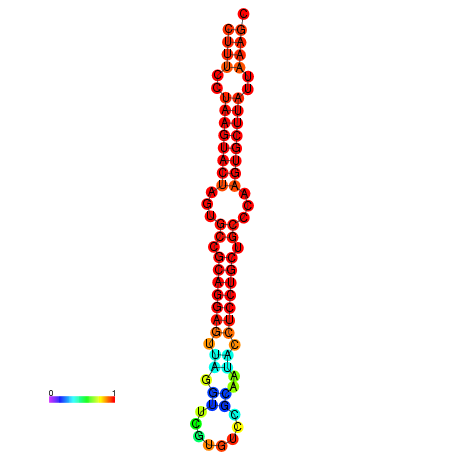 |
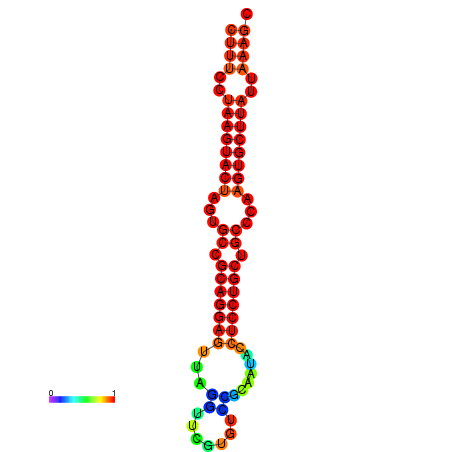 |
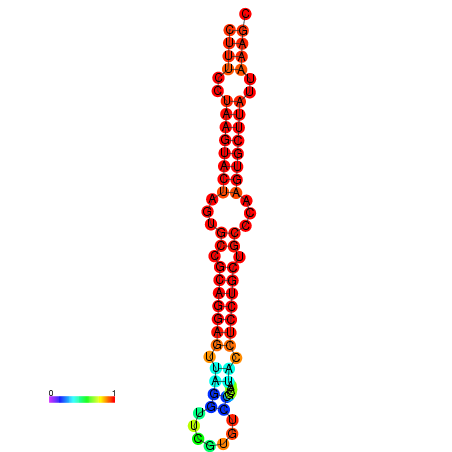 |
| dG=-25.7, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 |
|---|---|---|---|---|
 |
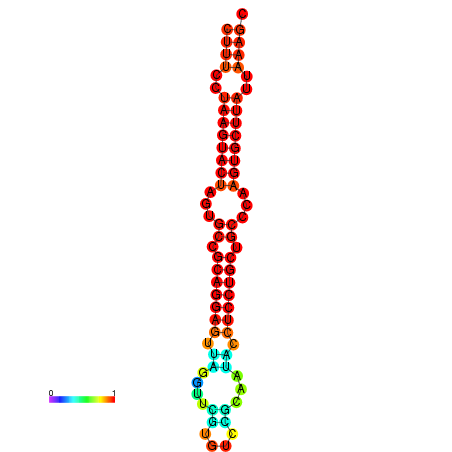 |
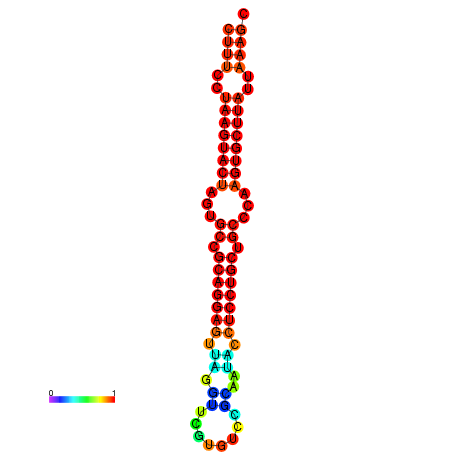 |
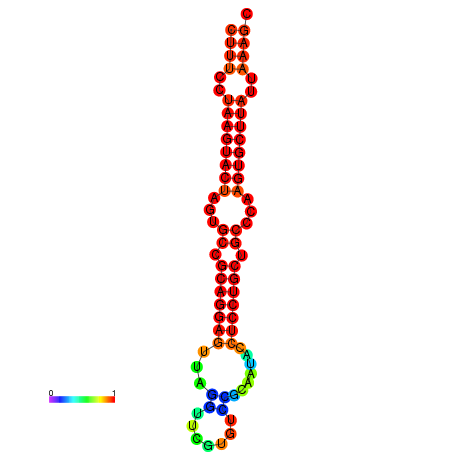 |
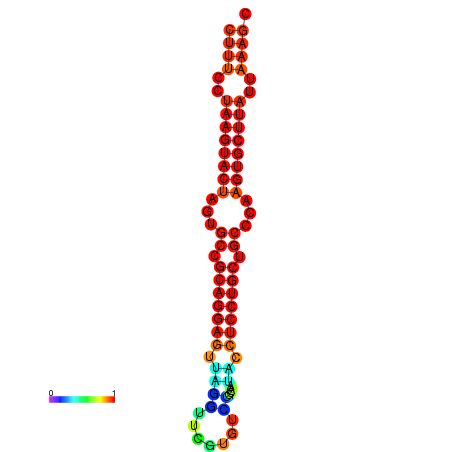 |
| dG=-25.7, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
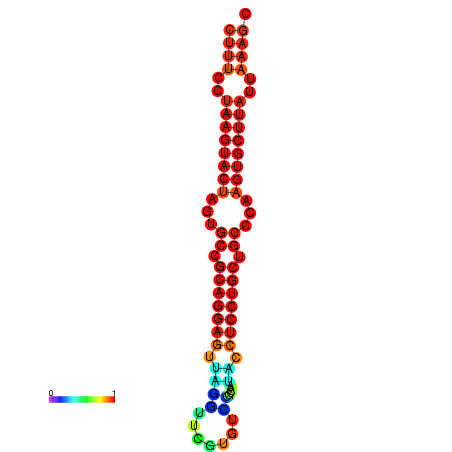 |
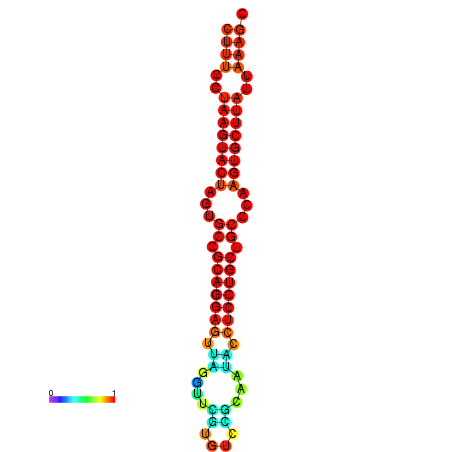
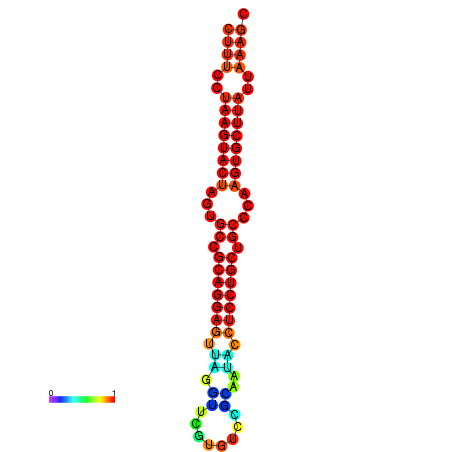
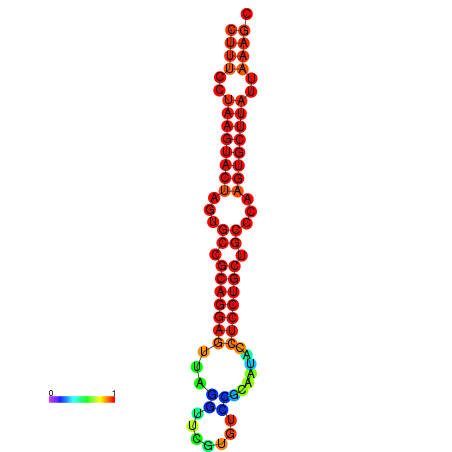
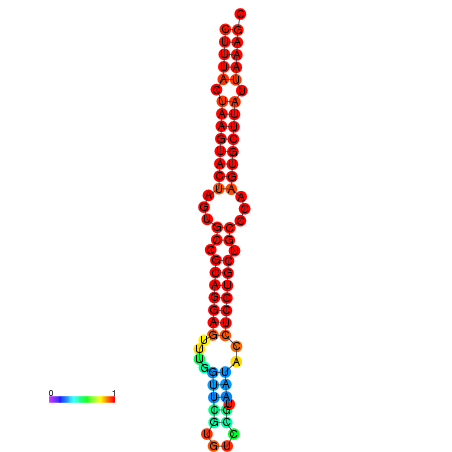
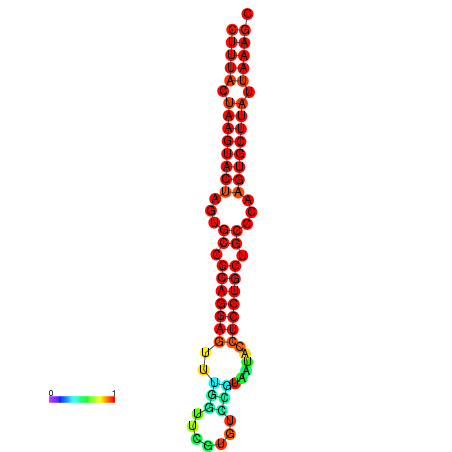
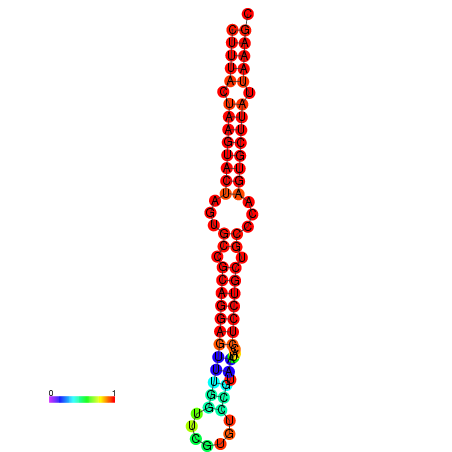
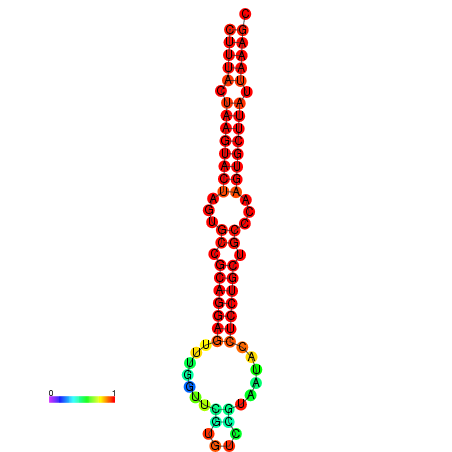
| dG=-27.4, p-value=0.009901 | dG=-27.3, p-value=0.009901 | dG=-26.8, p-value=0.009901 | dG=-26.8, p-value=0.009901 | dG=-26.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
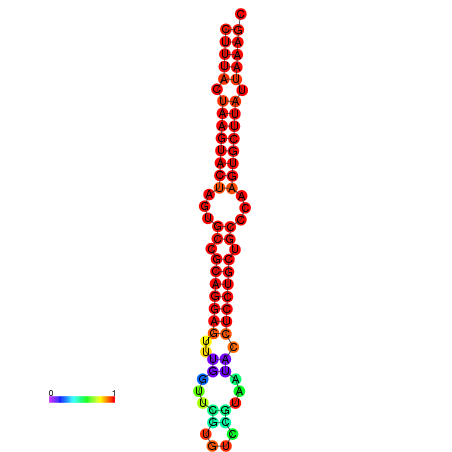 |
| dG=-26.7, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-26.0, p-value=0.009901 | dG=-26.0, p-value=0.009901 |
|---|---|---|---|---|
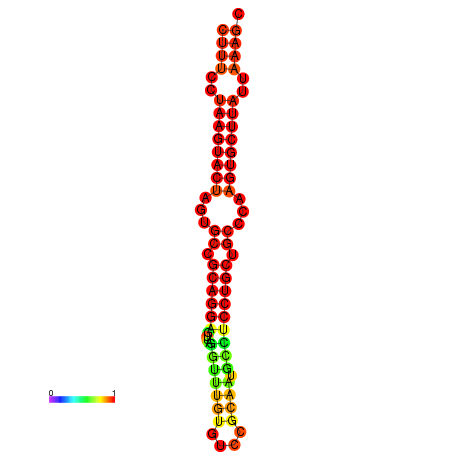 |
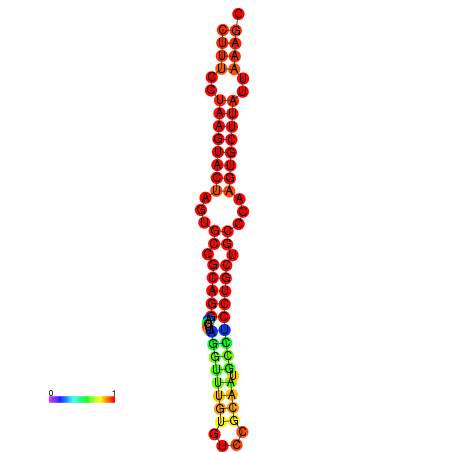 |
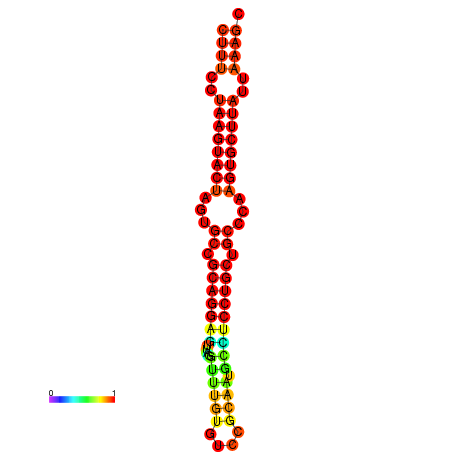 |
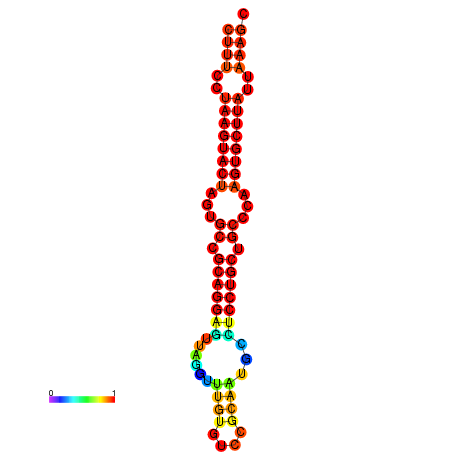 |
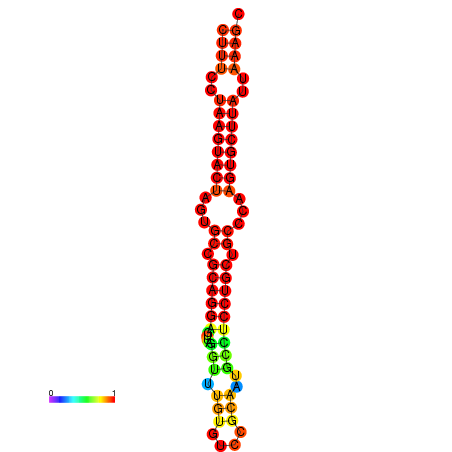 |
| dG=-27.8, p-value=0.009901 |
|---|
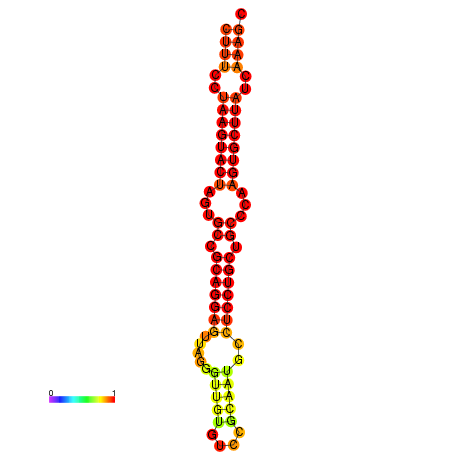 |
| dG=-26.4, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|---|---|
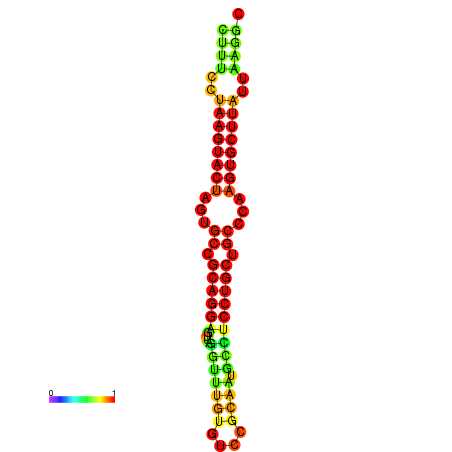 |
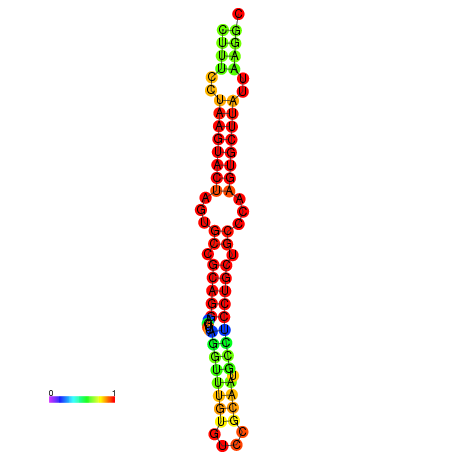 |
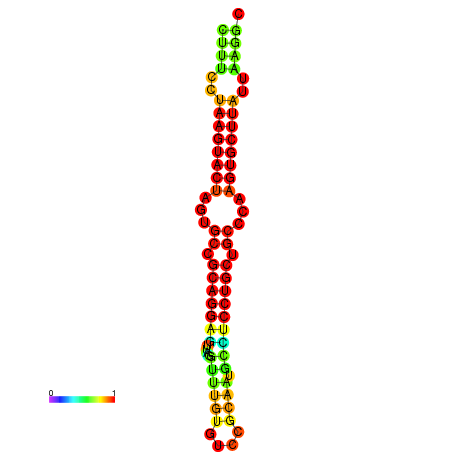 |
 |
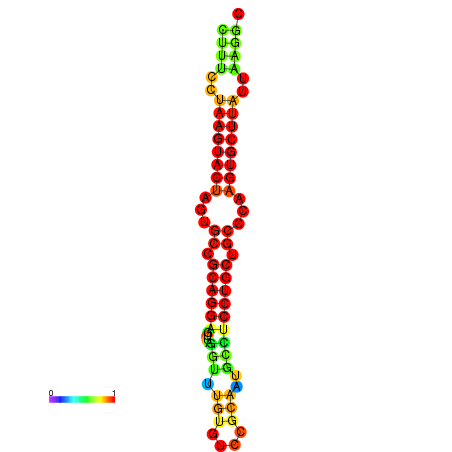 |
Generated: 03/07/2013 at 04:59 PM