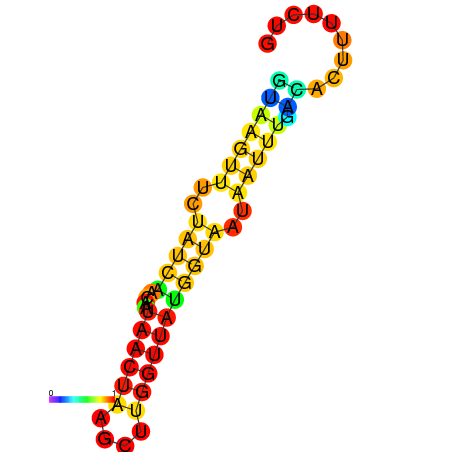
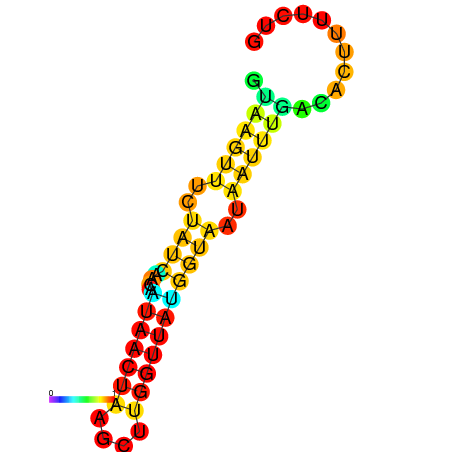
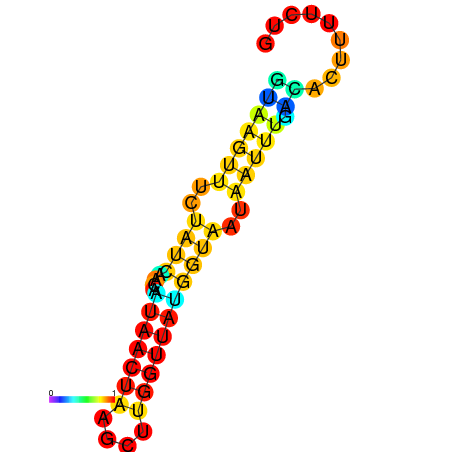
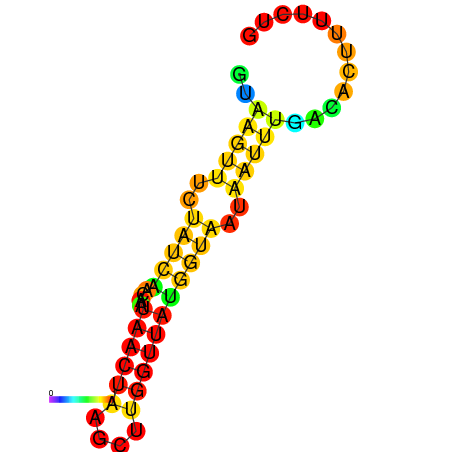
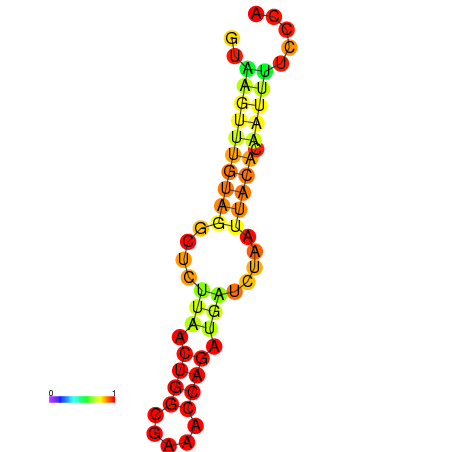
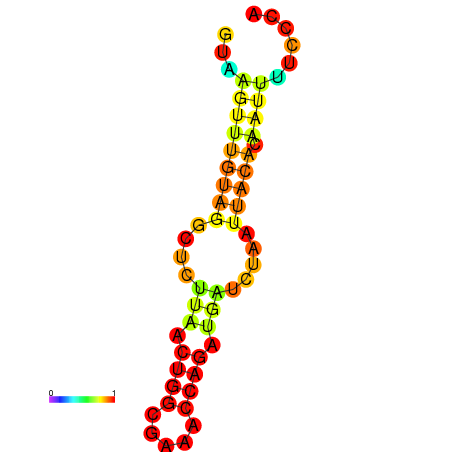
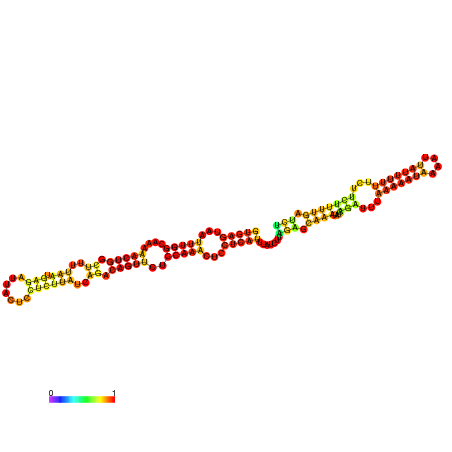
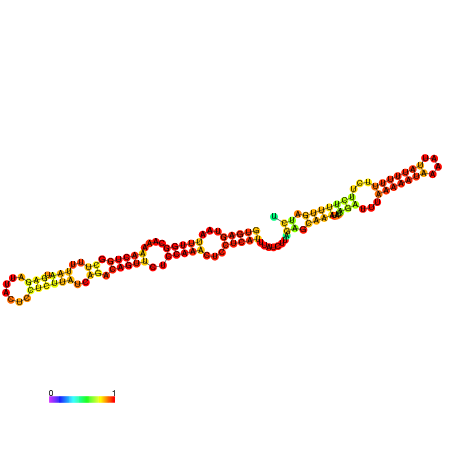
| dm3 |
chr3L:8104192-8104292 + |
GGTGGTAAAAGTGGAGTAAGTTTGGAGTG----CCTGGGCTAAGAACTTCCATT---------------------------------------------------------CTGAAACTTAACTCTTGCAATCAAACTTTC------------------------------------------------------------------TTTCAGAACCAAGCGTGGCAG |
| droSim1 |
chr3L:7572156-7572239 + |
GGTGGTAAAAGTGGAGTAAGTTTGGAGTC----CCTGGGCTAAGAACTTCCATT--------------------------------------------------------------------------CAATCAAACTTTC------------------------------------------------------------------TTTCAGAACCAAGCGTGGCAG |
| droSec1 |
super_0:391632-391715 + |
GGTGGTAAAAGTGGAGTAAGTTTGGAGTC----CCTGGGCTAAGAACTTCCATT--------------------------------------------------------------------------GAAGCAAACTTTC------------------------------------------------------------------TTTTAGAACCAAGCGTGGTAG |
| droYak2 |
chr3L:8691884-8691979 + |
CGTTGTAAAAGTGGAGTAAGTTTGCGA----------------------------------------------------------------CTTACGTTC-CTTTTATATACCG-ACATACCGTCTTACAACCAAACTTTC------------------------------------------------------------------TTTCAGAACTAAGCGCGGCAG |
| droEre2 |
scaffold_4784:3260723-3260819 - |
GGTGGTAAAAGTGGAGTAAGCTTGCAA----------------------------------------------------------------CTTGAGTTATATCTTTTATCCCG-CTATACTTACTTACAATCAAACTTTC------------------------------------------------------------------TCGCAGAACCAAGCGCGGCAG |
| droAna3 |
scaffold_13337:13543116-13543232 - |
AGTGGTTAAAGTGGAGTAAGTGGCGT----------------------TTAAGGG------AGTGATG----------G---ATAGATGGACCTGAGTCT-TTTTTAGT---TGTTCTCTTTCTCTAACATTCATAATCCG------------------------------------------------------------------TTTCAGAAACAAACGTGGCCG |
| dp4 |
chrXR_group3a:1131777-1131868 + |
GGTGGTCAAAGTAGAGTAAGTGATCC----------------------CCACCGA------ACTATTGTGACTTTACGC------T-------------C-TAATTGGG----------------------TACATTTTTT------------------------------------------------------------------GTGCAGAACCAAGCGTGGCCG |
| droPer1 |
super_23:1158678-1158769 + |
GGTGGTCAAAGTAGAGTAAGTGATCC----------------------CCACCGA------ACTATTGTGGCTTTACGC------T-------------C-TAATTGGG----------------------TACATTTTTT------------------------------------------------------------------GTGCAGAACCAAGCGTGGCCG |
| droWil1 |
scaffold_180727:72451-72556 + |
AGTGGTCAAAGTACAGTAAGTACATGG-----------AATGACCAATCCAATTAACTATA--TA-TGTACATATATGT------T-------------A-AATCCAAC----------------------CATCTCTTTT------------------------------------------------------------------ATCCAGAAATAGACGTGGCCG |
| droVir3 |
scaffold_13049:9356761-9356852 - |
GGTTGTCAAAGTTGAGTAAGTTTCTA----------------------TCAAC----AATAACTA-AGCTTGGTTATGG------TA------------A-TAATTTG------------------------ACACTTTTC------------------------------------------------------------------TGTTAGAAATAAGCGCGGTCG |
| droMoj3 |
scaffold_6680:11400964-11401055 + |
CGTTGTCAAGGTTGAGTAAGTTTGTAGGC----TCTTAACT-----------------------GGCGAAACCAGATGA------T-------------C-TAATTAC-----------------------ACAATTTTCC------------------------------------------------------------------CATTAGAAACAAACGTGGTCG |
| droGri2 |
scaffold_15110:6804388-6804550 + |
CGTTGTCAAAGTCGAGTGAGTAATTTGGCAAAAACTGGCTT-------TTAATG-------------------AGATTACTCC--T-------------C-TTATCAG-----------------------ACAGTTCTCCAAACTCCTCATTTATCTTAGAGCAAATATCAGATTTAAAAATAAAATTATTTTTTCTTCTTTTGATCTTCAGAAATAAGCGCGGTCG |