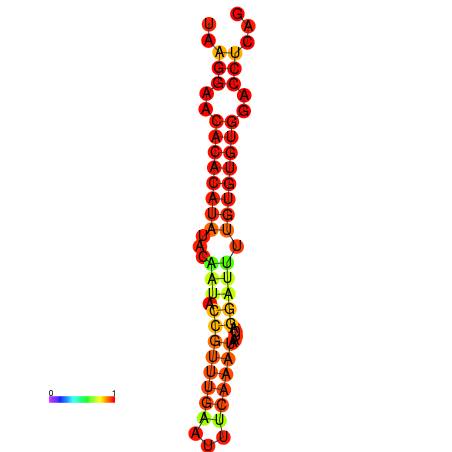
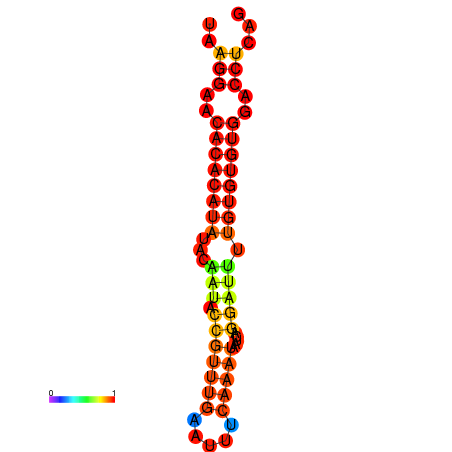
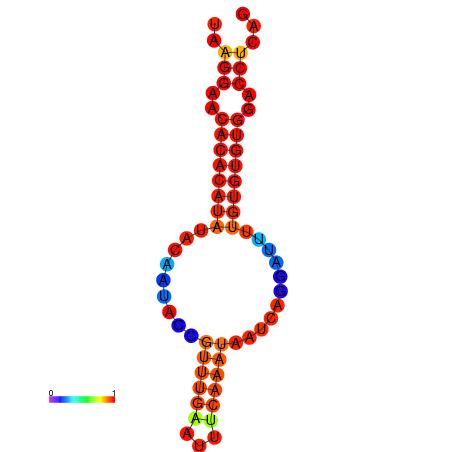
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:16991543-16991639 - | AAGAACTTTCTGCAGGTAAGGA----------------ACACACATATACAATACCG-------TTTGA-------------ATTTCAAATAATCAGGA------TTTTGTGTGTGGACCTCAGATCATGTTCCAGGCC |
| droSim1 | chr3L:16333274-16333368 - | AAGAACTTTCTGCAGGTAAGGA----------------ACACACAT--ACAGTCCCT-------GTTGA-------------ATTTCAAATAATCAGGA------TTTTGTGTGTGGACCTCAGATCATGTTCCAGGCC |
| droSec1 | super_0:9052557-9052651 - | AAGAACTTTCTGCAGGTAAGGA----------------ACACACAT--ACAATCCCT-------GTTGA-------------ATATTAAATAATCAGGA------TTTTGTGTGTGGACCTCAGATCATGTTCCAGGCC |
| droYak2 | chr3L:17082640-17082734 + | AAGAACTTTCTACAGGTCAGGA----------------TCACACAT--ACATTCCCT-------GTAGA-------------ATTTAATTTAATCAGGA------ATTTATGTGTGAACTTTAGATCATGTTCCAGGCC |
| droEre2 | scaffold_4784:16851458-16851552 + | AAGAATTTTCTGCAGGTAAGGA----------------TCGCACAT--ACAATCCCT-------GTCGC-------------ATTTTATGTAATCAGGA------TTTTGCGTGTGAACTTTAGATCATGTTCCAGGCC |
| droAna3 | scaffold_13337:13625775-13625865 + | AAAAACTTTCTGCAGGTGAGCA----------------GTTCA----------------AGCTTAGCGAGGTCTGTTCCAGTT-TCCTAACATTC----------TTCCGCTC-----CTTTAGATCATGTTCCAGGCC |
| dp4 | chrXR_group8:4827167-4827262 - | AAGAACTTTTTGCAGGTAAGCC----ATCACC------------AGCA-------TTGAAAGCCATTGA-------------AATGCGTCTAAATAGCAGTCT--CTCTCCTC-----CCACAGATCATGTTCCAGGCC |
| droPer1 | gnl|ti|780630171:515-611 - | AAGAACTTTTTGCAGGTAAGCC----ATCACC------------AGCA-------TTGAAAGCCATTGA-------------AATGCGTCTAAATAGCAGTCT--CTCTCCTC-----CCACAGATCATGTTCCAGGCC |
| droWil1 | scaffold_180955:197087-197182 - | AAAAACTTTTTACAGGTAAGCC----------------ACAACGAT--GAATGGCTCGAGGGAGATTGA-------------ACTCTATGTAATC-------TCTTCACTTTC-----TTGCAGATAATGTTTCAGGCT |
| droVir3 | scaffold_13049:10166522-10166615 + | AAGAACTTTCTGCAGGTGCGTT----CCCCCTGGCTGATCACATAT--GTCGTTT---------------------------ATACTATATTTT-CGCC------TTTTGCTC-----CTGCAGATCATGTTCCAGGCC |
| droMoj3 | scaffold_6680:10629193-10629280 - | AAGAACTTTCTCCAGGTGAGTT----ACGG-------TTCAC------ATAATTC----------TCGT-------------ATTCCAAATAACGAAAT------CTTTGTCC-----TTGCAGATCATGTTCCAGGCC |
| droGri2 | scaffold_15110:9071735-9071831 - | AAGAACTTGCTGCAGGTGAGATTTTGATCG-------ATTAA------TTAAGTC----------TGG--------CTCTCTTTGCTAAATATGCC--A--CT--TTTTGTGT-----TGGCAGATCATGTTCCAGGCC |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:16991543-16991639 - | AAGAACTTTCTGCAGGTAAGGAACACACATATACAATACCGTTTGAATTTCAAATAATCAGGATTTTGTGTGTGGACCTCAGATCATGTTCCAGGCC |
| droSim1 | chr3L:16333274-16333368 - | AAGAACTTTCTGCAGGTAAGGAACACACA--TACAGTCCCTGTTGAATTTCAAATAATCAGGATTTTGTGTGTGGACCTCAGATCATGTTCCAGGCC |
| droSec1 | super_0:9052557-9052651 - | AAGAACTTTCTGCAGGTAAGGAACACACA--TACAATCCCTGTTGAATATTAAATAATCAGGATTTTGTGTGTGGACCTCAGATCATGTTCCAGGCC |
| droYak2 | chr3L:17082640-17082734 + | AAGAACTTTCTACAGGTCAGGATCACACA--TACATTCCCTGTAGAATTTAATTTAATCAGGAATTTATGTGTGAACTTTAGATCATGTTCCAGGCC |
| droEre2 | scaffold_4784:16851458-16851552 + | AAGAATTTTCTGCAGGTAAGGATCGCACA--TACAATCCCTGTCGCATTTTATGTAATCAGGATTTTGCGTGTGAACTTTAGATCATGTTCCAGGCC |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-17.0, p-value=0.009901 | dG=-16.3, p-value=0.009901 | dG=-16.0, p-value=0.009901 |
|---|---|---|
 |
 |
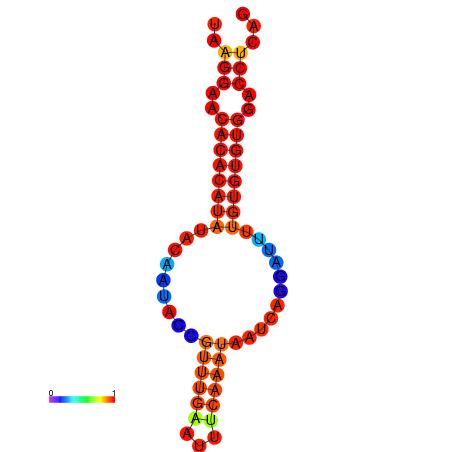 |
| dG=-20.7, p-value=0.009901 | dG=-20.7, p-value=0.009901 | dG=-20.7, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 |
|---|---|---|---|---|
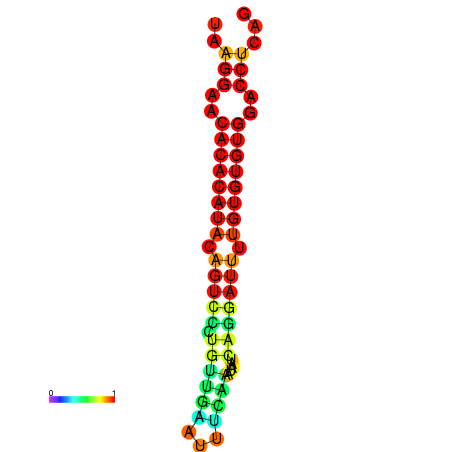 |
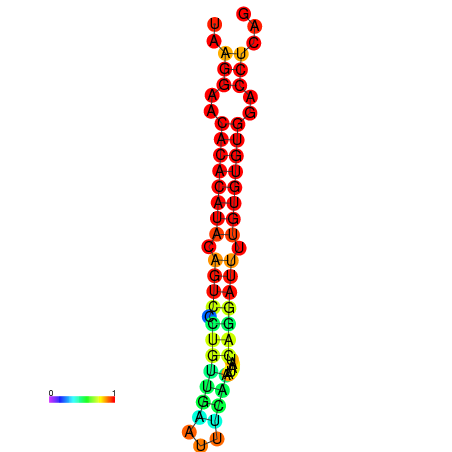 |
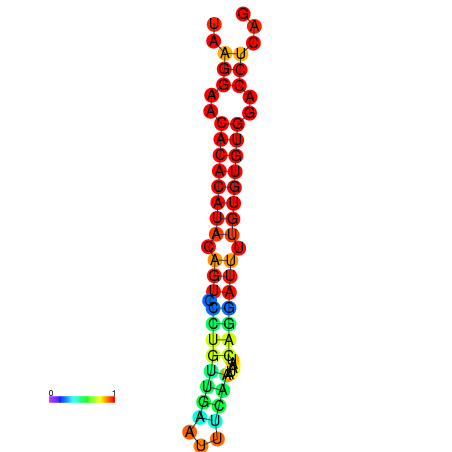 |
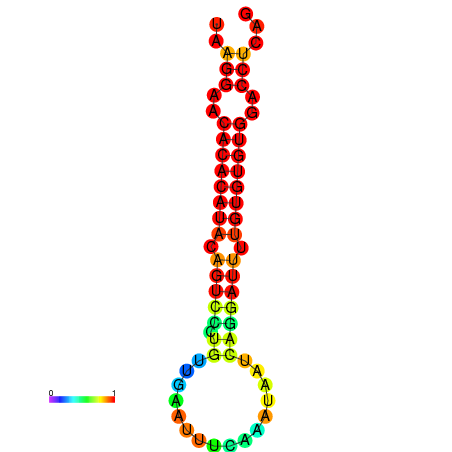 |
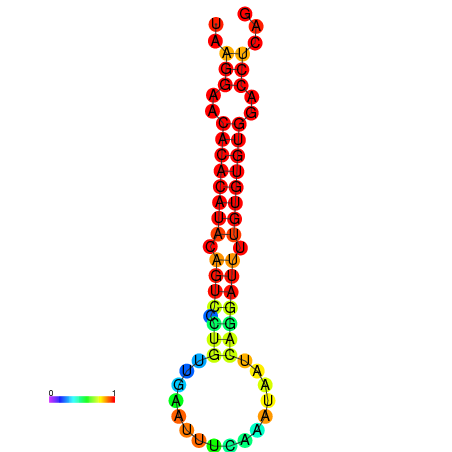 |
| dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-20.1, p-value=0.009901 |
|---|---|---|---|---|
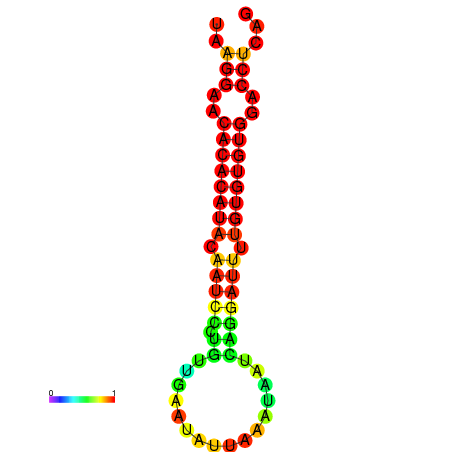 |
 |
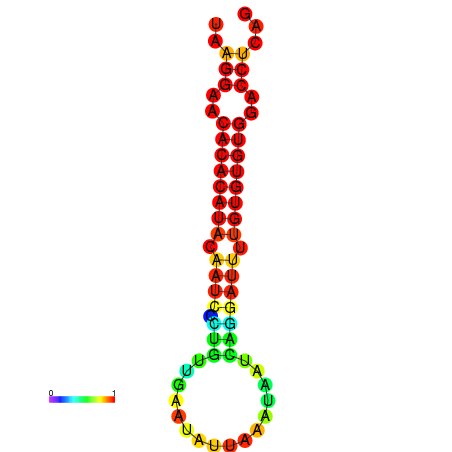 |
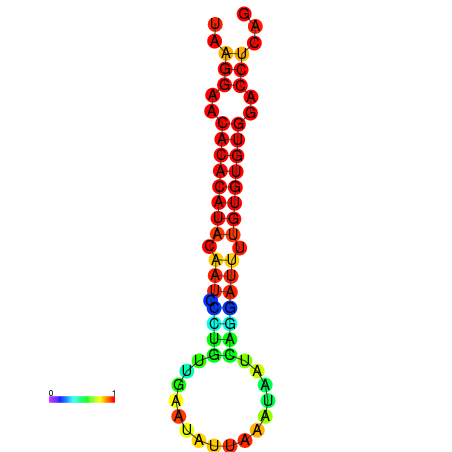 |
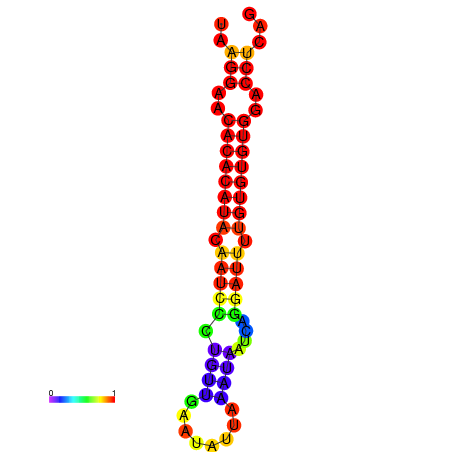 |
| dG=-18.8, p-value=0.009901 | dG=-18.8, p-value=0.009901 | dG=-18.8, p-value=0.009901 | dG=-18.5, p-value=0.009901 | dG=-18.5, p-value=0.009901 |
|---|---|---|---|---|
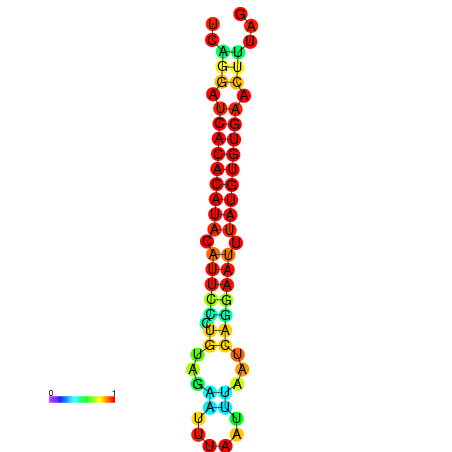 |
 |
 |
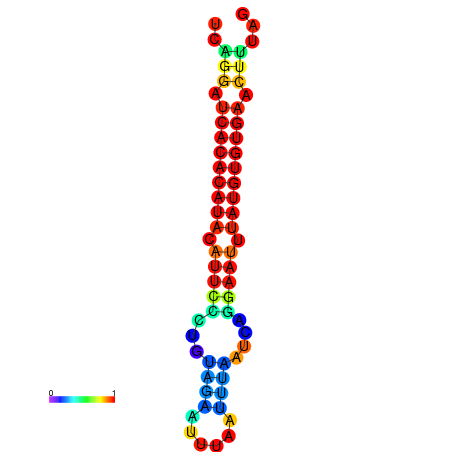 |
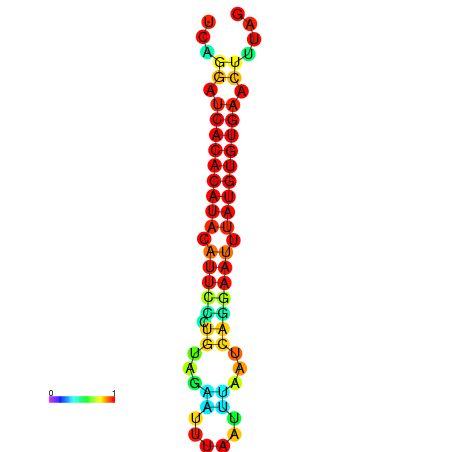 |
| dG=-18.2, p-value=0.009901 | dG=-18.0, p-value=0.009901 | dG=-18.0, p-value=0.009901 | dG=-18.0, p-value=0.009901 | dG=-17.4, p-value=0.009901 |
|---|---|---|---|---|
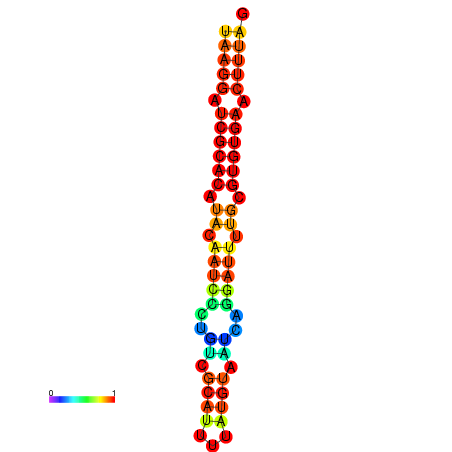 |
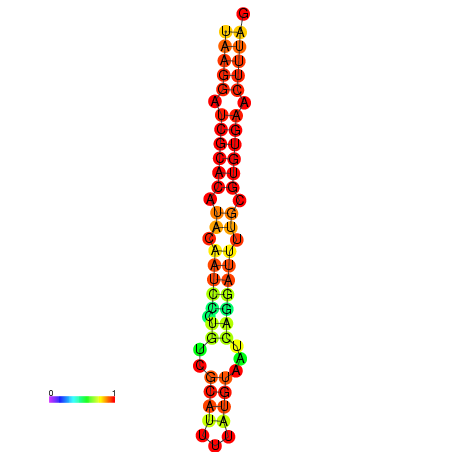 |
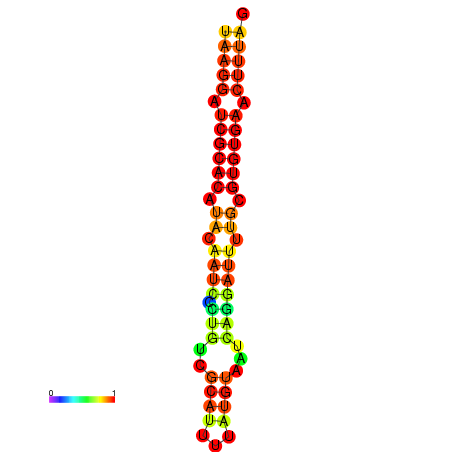 |
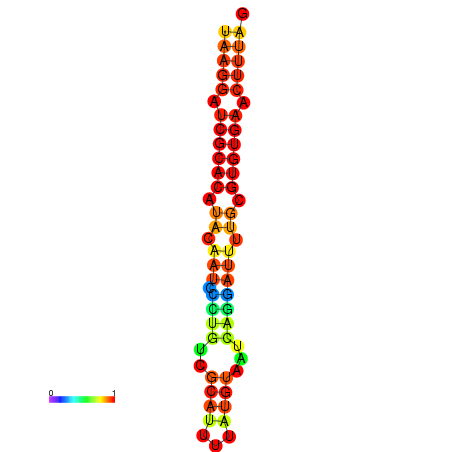 |
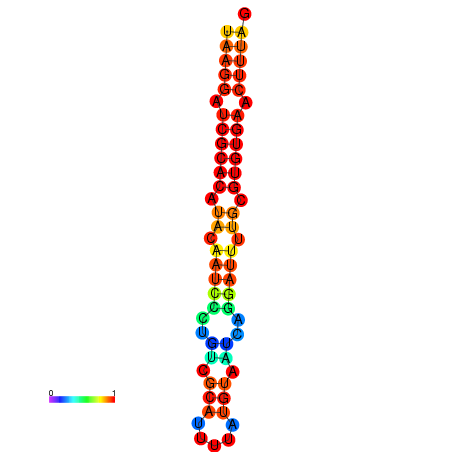 |
| dG=-10.1, p-value=0.009901 | dG=-10.1, p-value=0.009901 | dG=-9.9, p-value=0.009901 | dG=-9.9, p-value=0.009901 | dG=-9.9, p-value=0.009901 | dG=-9.9, p-value=0.009901 |
|---|---|---|---|---|---|
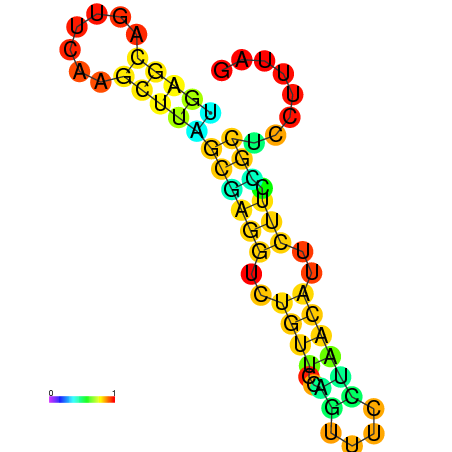 |
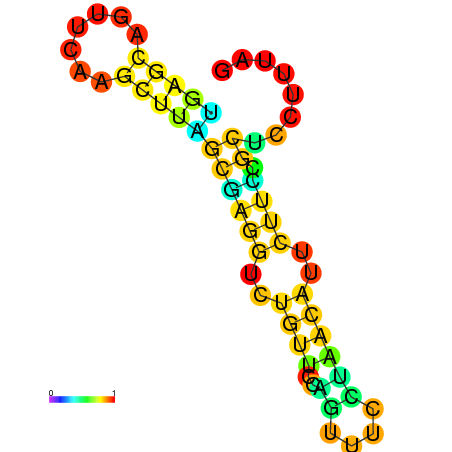 |
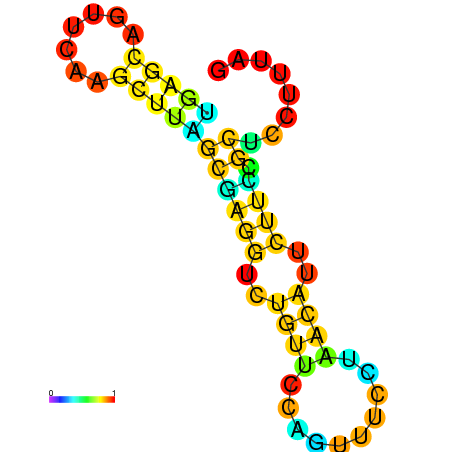 |
 |
 |
 |
| dG=-5.1, p-value=0.009901 | dG=-4.8, p-value=0.009901 | dG=-4.6, p-value=0.009901 | dG=-4.1, p-value=0.009901 |
|---|---|---|---|
 |
 |
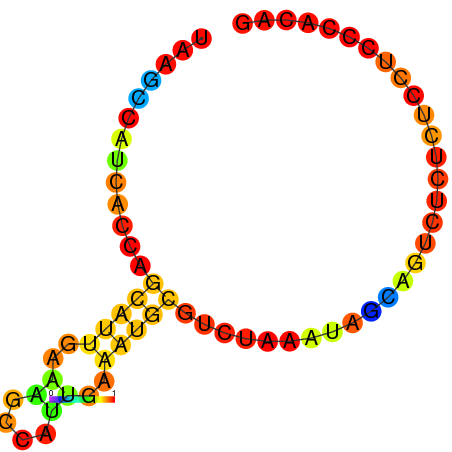 |
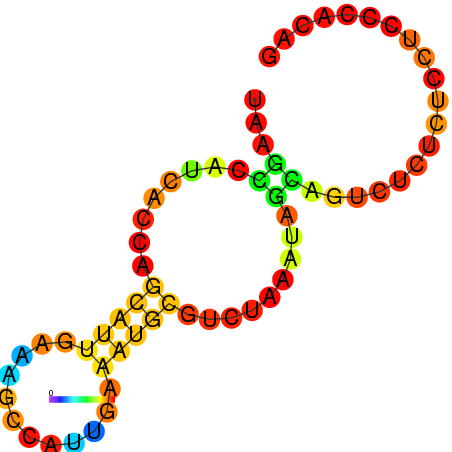 |
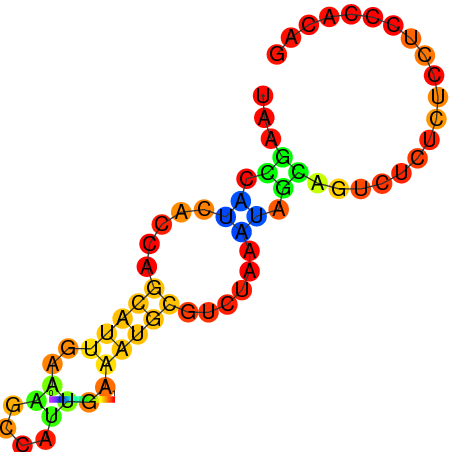
| dG=-5.1, p-value=0.009901 | dG=-4.8, p-value=0.009901 | dG=-4.6, p-value=0.009901 | dG=-4.1, p-value=0.009901 |
|---|---|---|---|
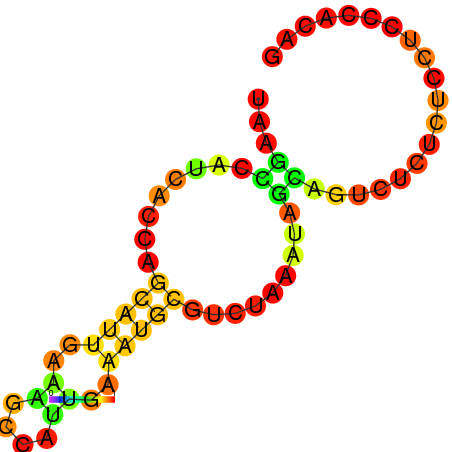 |
 |
 |
 |
| dG=-12.0, p-value=0.009901 | dG=-12.0, p-value=0.009901 | dG=-11.9, p-value=0.009901 | dG=-11.9, p-value=0.009901 | dG=-11.9, p-value=0.009901 | dG=-8.4, p-value=0.009901 |
|---|---|---|---|---|---|
 |
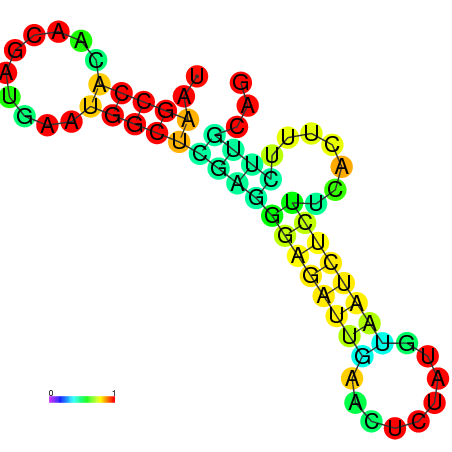 |
 |
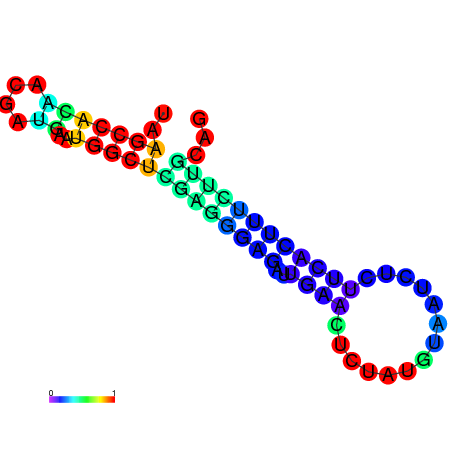 |
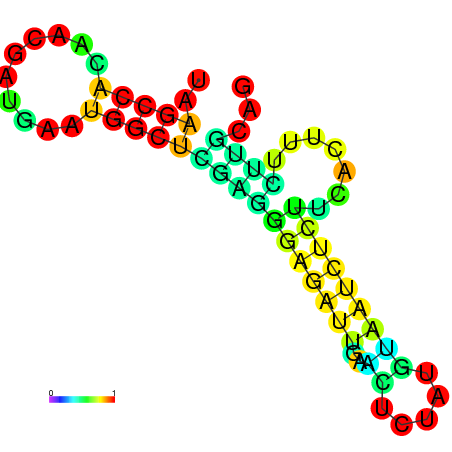 |
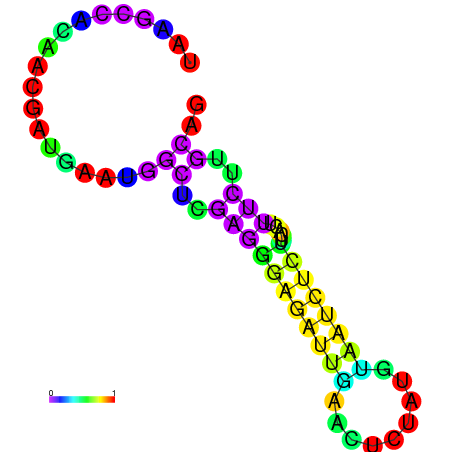 |
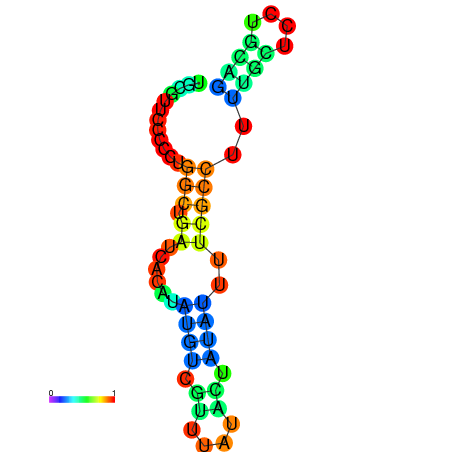
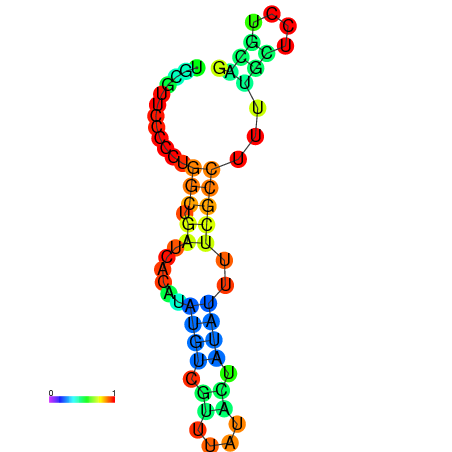
| dG=-5.9, p-value=0.009901 | dG=-5.7, p-value=0.009901 | dG=-5.6, p-value=0.009901 | dG=-5.5, p-value=0.009901 | dG=-5.4, p-value=0.009901 |
|---|---|---|---|---|
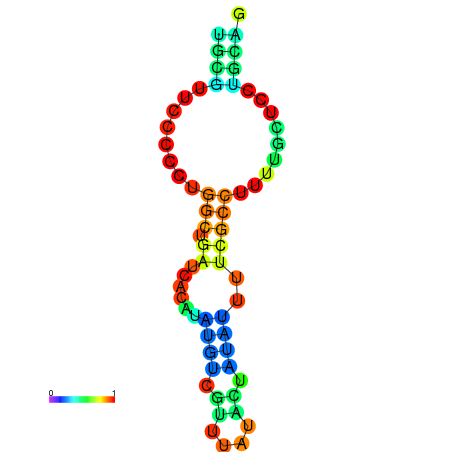 |
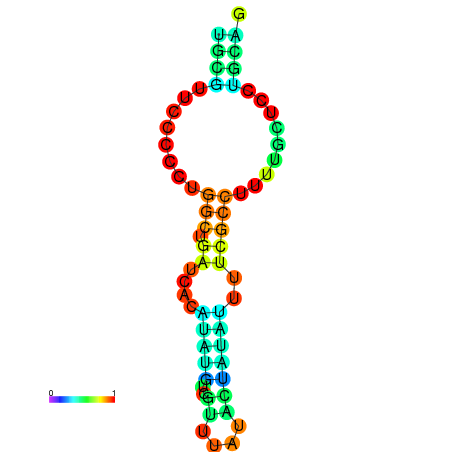 |
 |
 |
 |
| dG=-3.0, p-value=0.009901 | dG=-3.0, p-value=0.009901 | dG=-2.8, p-value=0.009901 | dG=-2.8, p-value=0.009901 | dG=-2.6, p-value=0.009901 | dG=-2.6, p-value=0.009901 |
|---|---|---|---|---|---|
 |
 |
 |
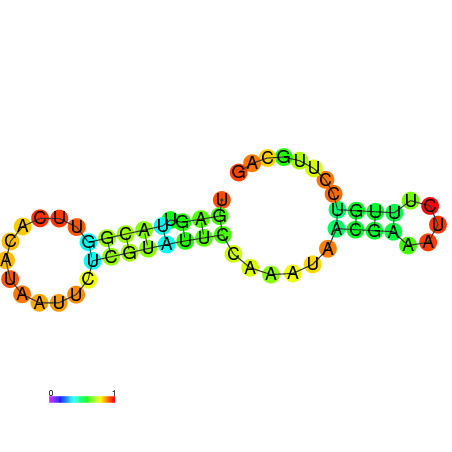 |
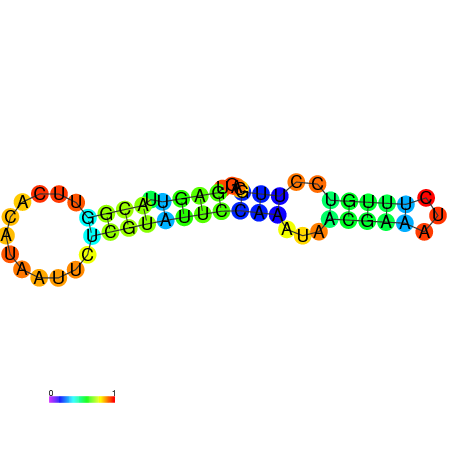 |
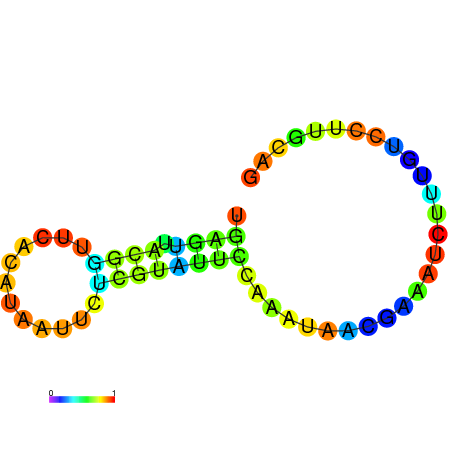 |
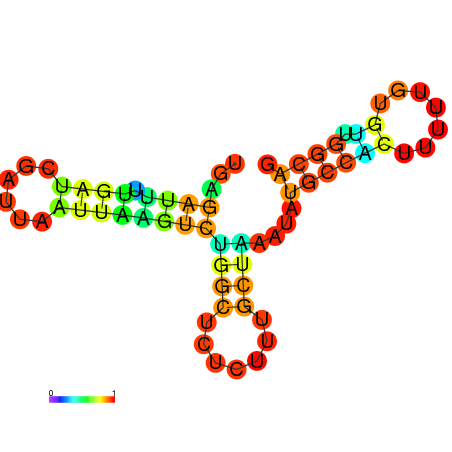
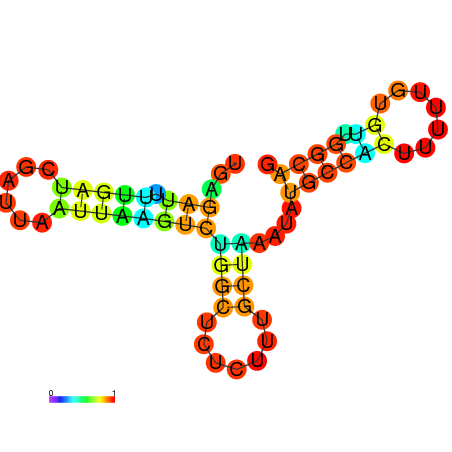
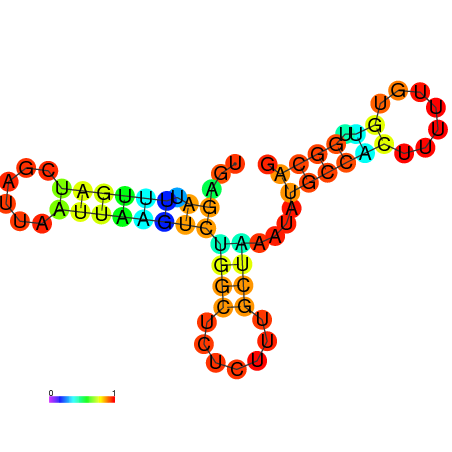
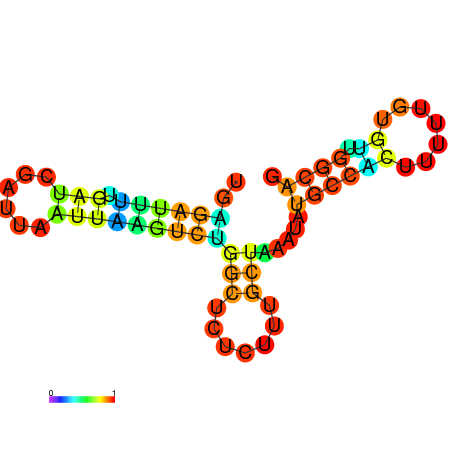
| dG=-10.0, p-value=0.009901 | dG=-10.0, p-value=0.009901 | dG=-10.0, p-value=0.009901 | dG=-10.0, p-value=0.009901 | dG=-9.8, p-value=0.009901 | dG=-7.7, p-value=0.009901 |
|---|---|---|---|---|---|
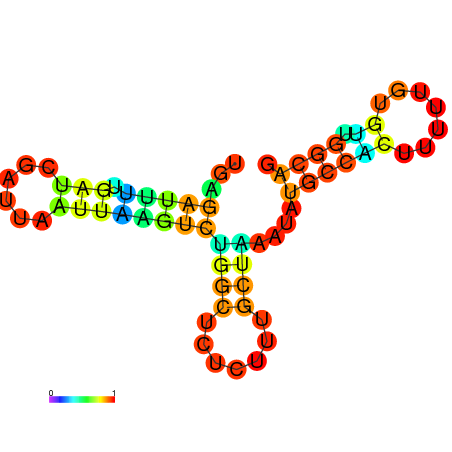 |
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 04:58 PM