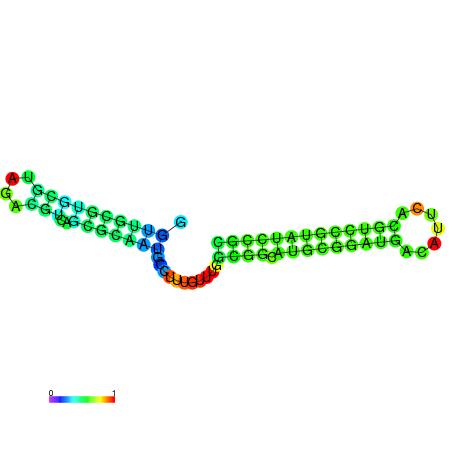
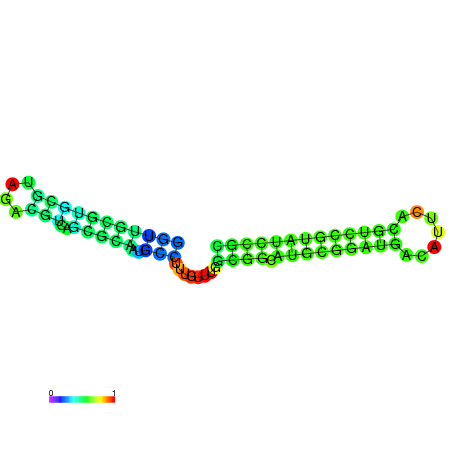
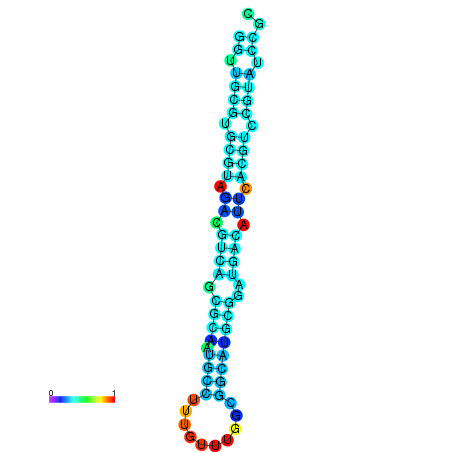
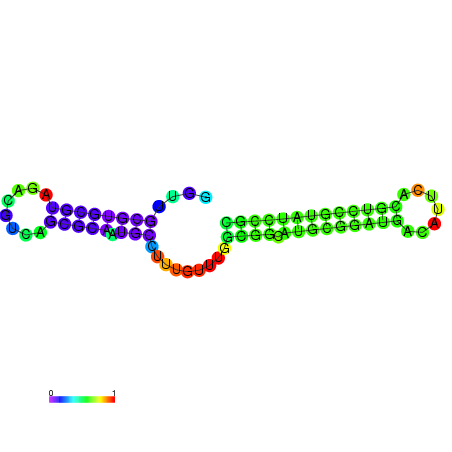
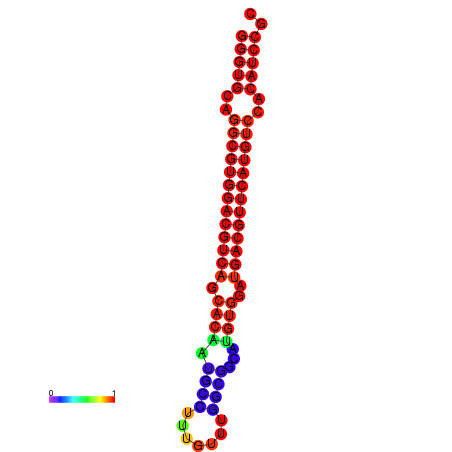
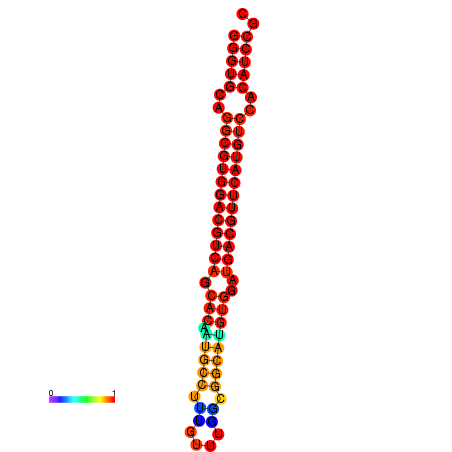
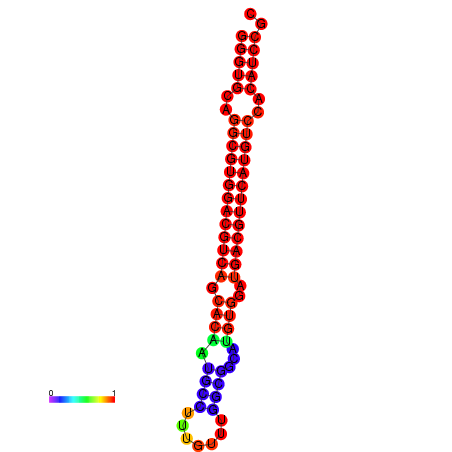
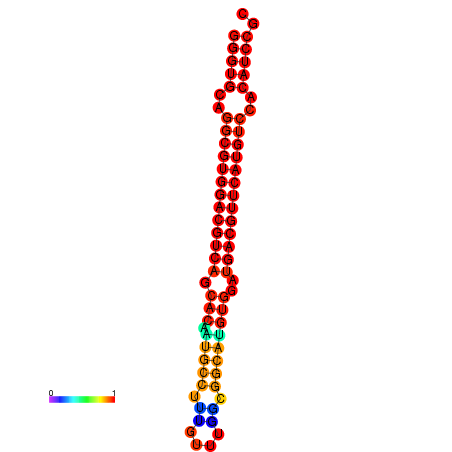
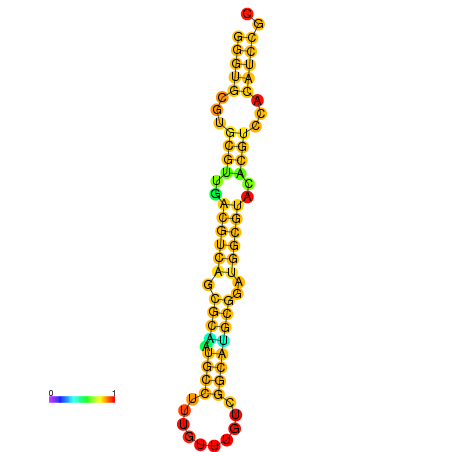
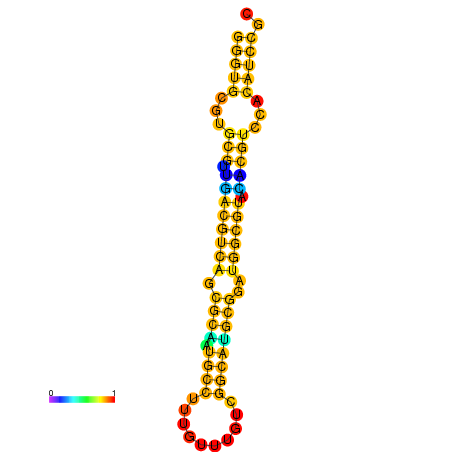
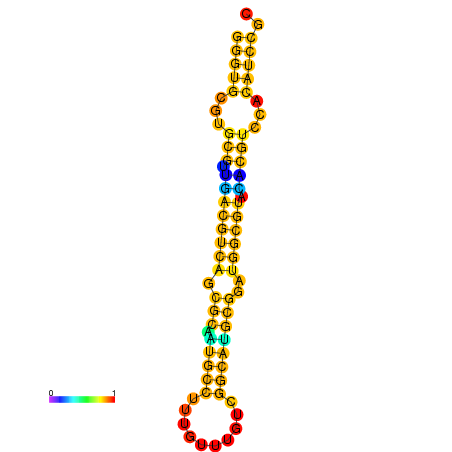
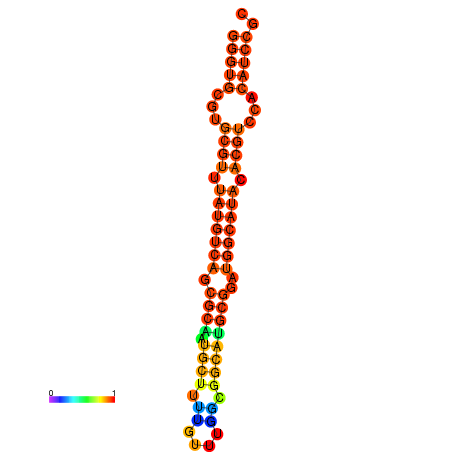
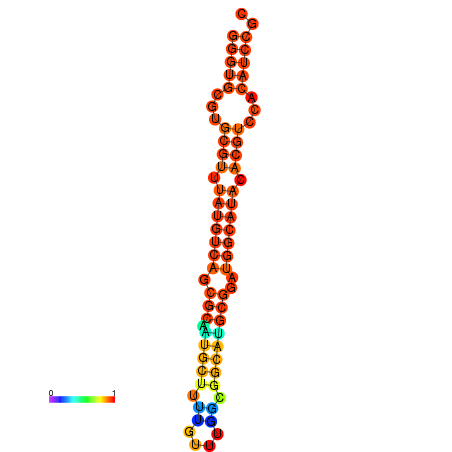
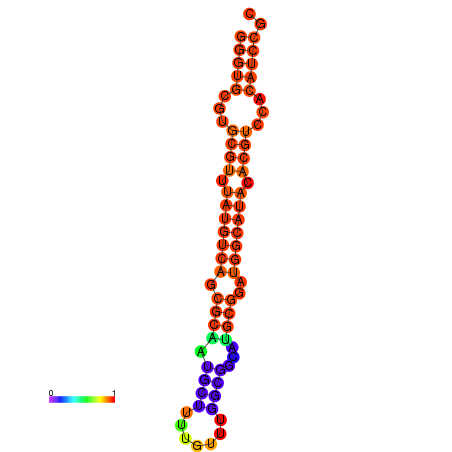
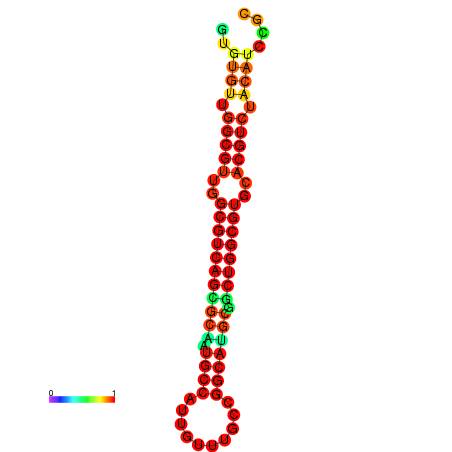
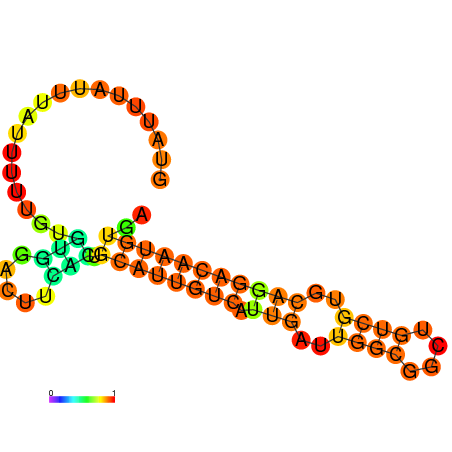
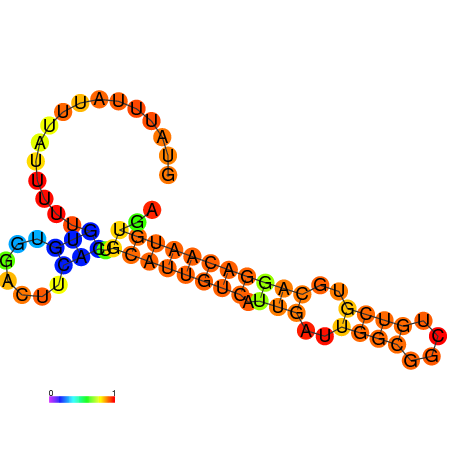
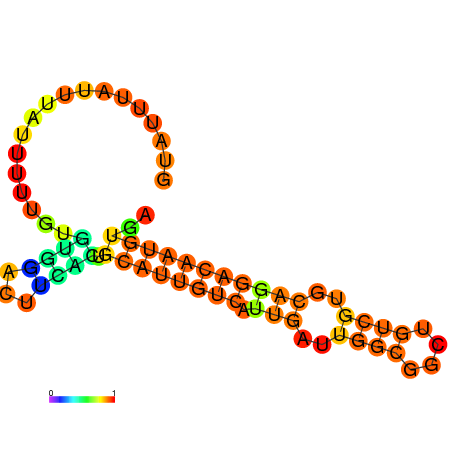
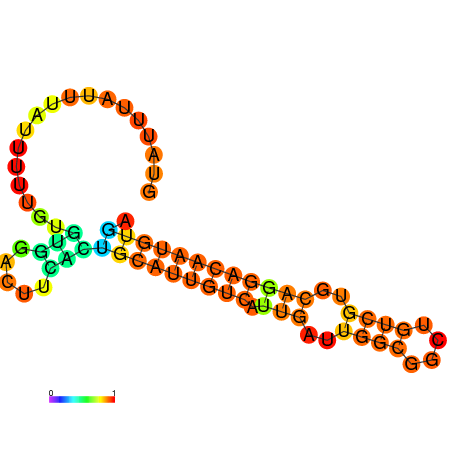
| dm3 |
chrX:19451026-19451151 + |
CTTTGGGCGTTCGCCTCCTTGCTGT------GGGTGTGGT--------TGCGTGCGTAGACGTCAGCGCAATGCCTTTGTTTGGCGGCATGCG-G---ATGACAT-----------------TCACGTCCGTATCCGCACACACATCCGCATCCATATA-----------------------------TC------ |
| droSim1 |
chrX:15016847-15016968 + |
CTTTGGGCGTTCGCCTCCTTGCTGT------GGG--TGGG--------TGCAGGCGTGGACGTCAGCACAATGCCTTTGTTTGGCGGCATGTG-G---ATGACGT-----------------TCATGTCCACATCCGCACACACATCCGCATCCATA-------------------------------TC------ |
| droSec1 |
super_8:1733318-1733439 + |
CTTTGGGCGTTCGCCTCCGTGCTGT------GGG--TGGG--------TGCAGGCGTGGACGTCAGCACAATGCCTTTGTTTGGCGGCATGTG-G---ATGACGT-----------------TCATGTCCACATCCGCACACACATCCGCATCCATA-------------------------------TC------ |
| droYak2 |
chrX:11887438-11887559 - |
CTTTGGGCGCTCTCCTCCTTGCTGT------GAG--CGGG--------TGCGTGCGTTGACGTCAGCGCAATGCCTTTGTTTGTCGGCATGCG-G---ATGGCGT-----------------ACACGTCCACATCCGCACACACACACGCATCTGTA-------------------------------TC------ |
| droEre2 |
scaffold_4690:9687367-9687488 + |
CTTTGGGCGCTCCTTTCCTGGCTGT------GAG--TGGG--------TGCGTGCGTTTATGTCAGCGCAATGCTTTTGTTTGGCGGCATGCG-G---ATGGCAT-----------------ACACGTCCACATCCGCACACACATCCGCATCCGTA-------------------------------TC------ |
| droAna3 |
scaffold_13417:2469661-2469798 - |
CGA-----------CTT----CTGTGTGT--GTGTGCGTG--------TGTTGGCGTTGGCGTCAGCGCAATGCCATTGTTTGCCGGCATGCG-G---CTGGCGT-----------------GCACGTCTACATCCGCACCTGCATCCACAT------AGCAGCTGCAACAACAAACACGGCACTCCGTC------ |
| dp4 |
chrXL_group1e:12520639-12520746 - |
TTTTTGA-------------GT-----GTGTGTT--TGTATTTATTTATTTTTGTGTGGACTTCACTGCATTGTCATTGATTGGCGGC-----------TGTCGT-----------------GCAGGACAATGT--GAAACATCATTCGACA-CAAA-------------------------------TC------ |
| droPer1 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_13042:909712-909778 - |
A----------------------------------------------------------------------GGCCATTGTT--ATGGCAGGTGGGCAGGTGTCAT-----------------CAATAGCCGCATCC--AGGCACATCCGCATCCACA-------------------------------T------- |
| droMoj3 |
scaffold_6328:2902676-2902778 - |
CTGTGGGCTTTAGGCCAATTGATAT------GGT--CGTG--------T------------------------------------GGCATGAG-G---CAGGTGTCAGCAGGTGGCAGCAATTCGGAACCAT-TGTGTGCCCACATCCGCATTTGCA-A-----------------------------GC------ |
| droGri2 |
scaffold_15081:2046312-2046418 - |
TGTTGCA-------------GTTGC------ATT--TGAA--------TGTAGGTGTTGTCATTA-------------TTTTGAGGGCATGCG-G---CAGGTGTCAACG------------CCACGTCCTCATTTGCATCCGCTTCCGCATCCGCA-------------------------------TCCGCTGT |