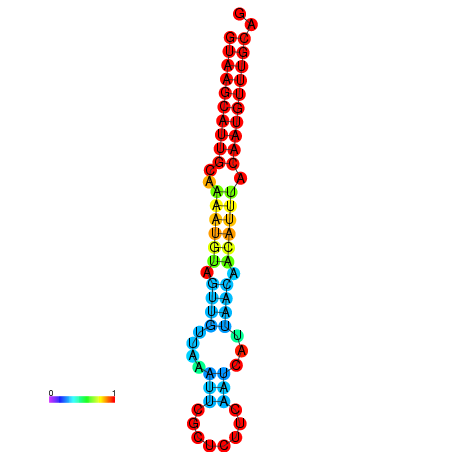
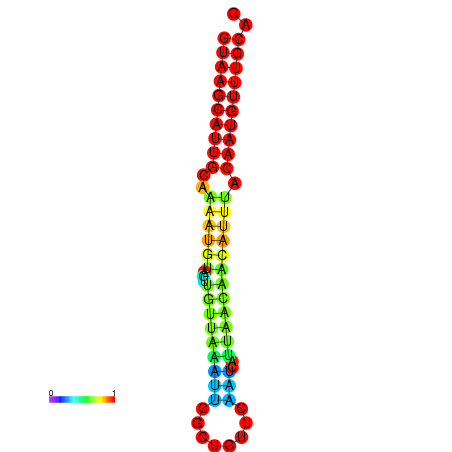
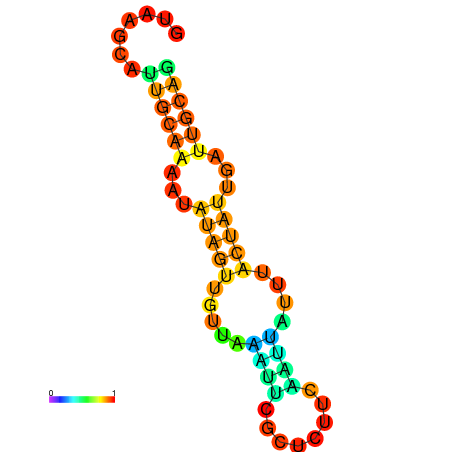
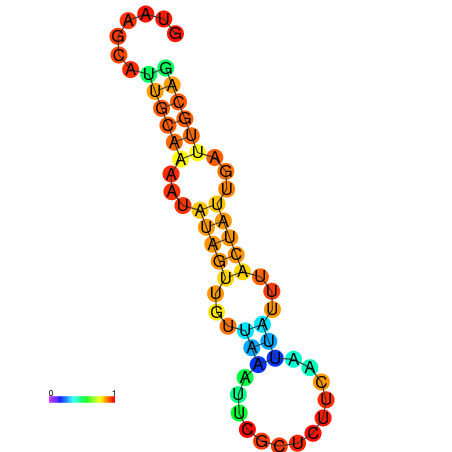
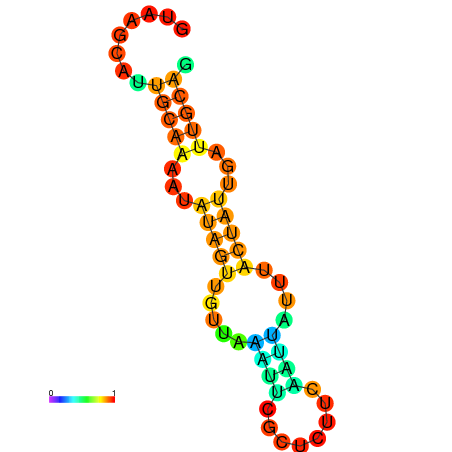
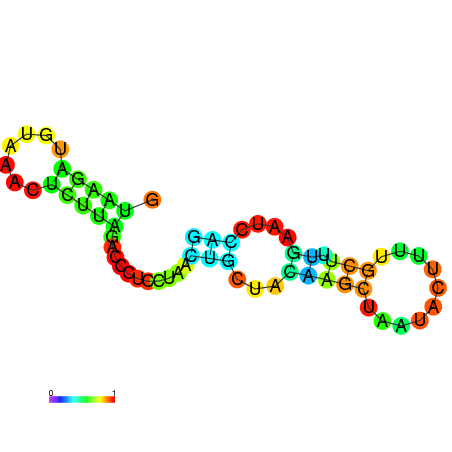
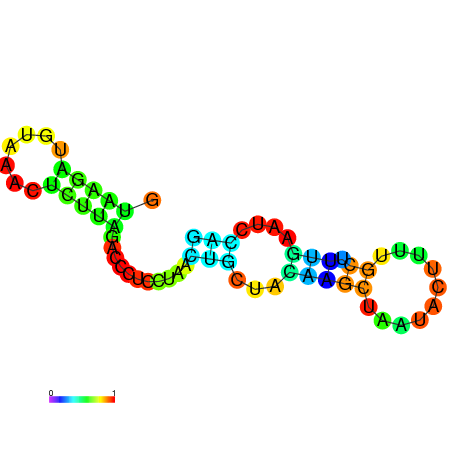
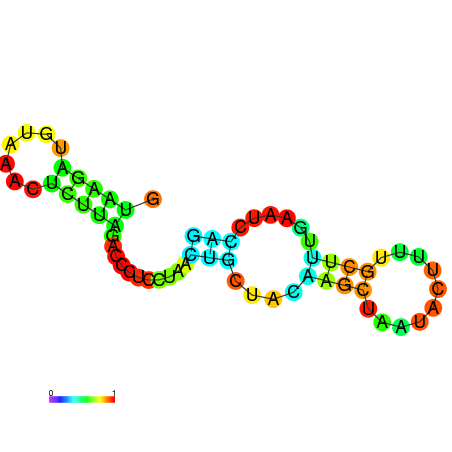
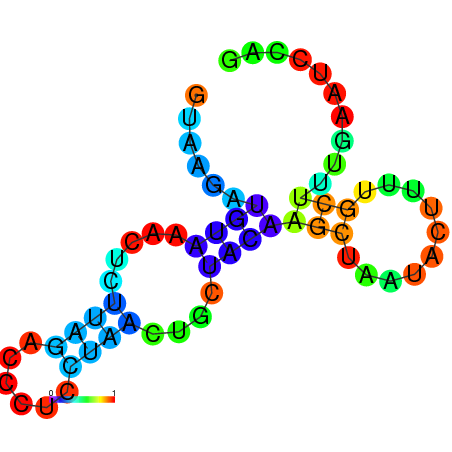
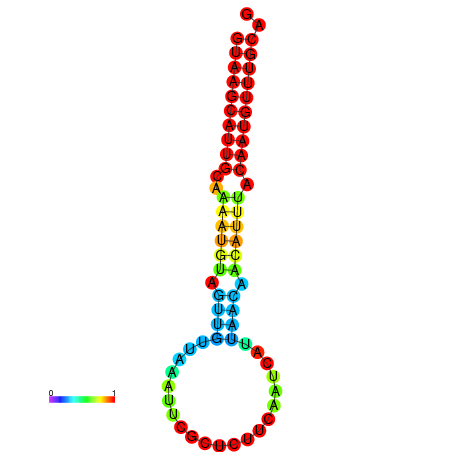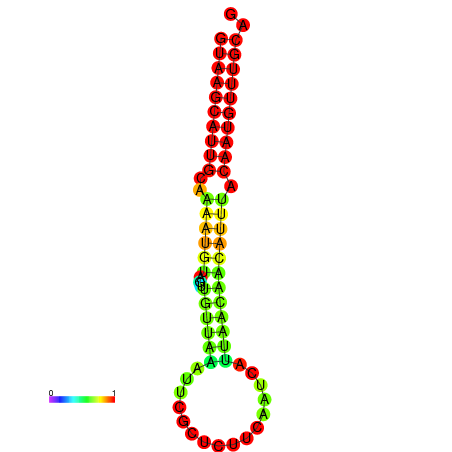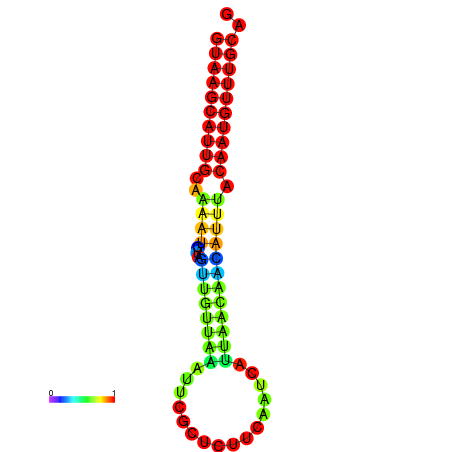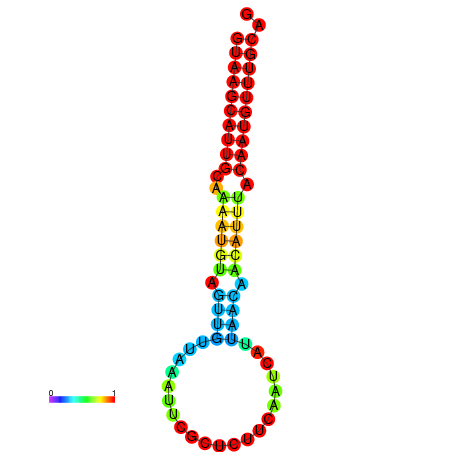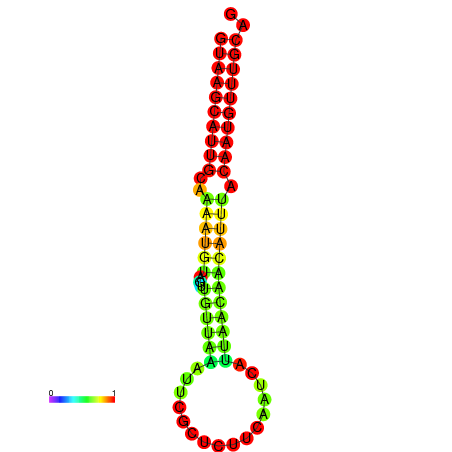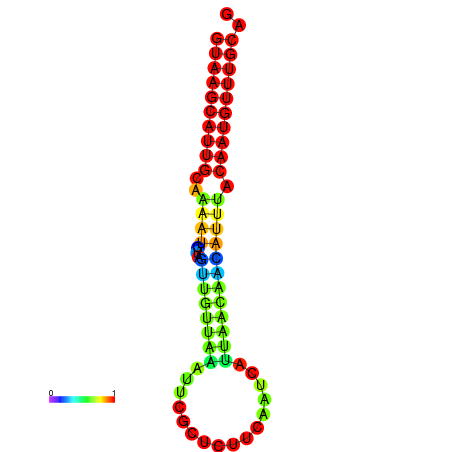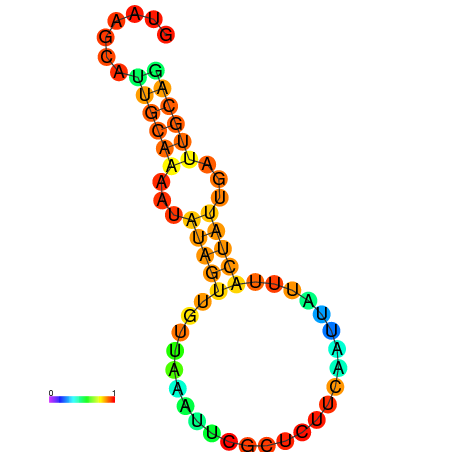| dm3 |
chr2L:16678235-16678332 + |
GATAAGCACGTCATCGTAAGCATTGCAAAATG--TAGTTGTTAAA-------TTCGC--------------TCTTCAATCATTA-ACAACATTT-----ACAATGTTTGCAGGCTGTCAACGTTGAT |
| droSim1 |
chr2L:16366233-16366322 + |
GATAAGCACGTCATCGTAAGCATTGCAAAATA--TAGTTGTTAAA-------TTCGC--------------TCTTCAATTATTT---------A-----CTATTGATTGCAGGCTGTCAACGTTGAT |
| droSec1 |
super_7:325110-325199 + |
GATAAGCATGTTATCGTAAGCATTGCAAAATA--TAGTTCTTAAA-------TTCGC--------------TCTTCAATTATTT---------A-----CTATTGATTGCAGGCTGTCAACGTTGAT |
| droYak2 |
chr2R:4424626-4424717 - |
GATAAGCACGTCATCGTAAGCTTTGCTATATATGTAGATGTGAAT-------TTCGA--------------GCTTCAATTCTTT---------T-----CTATTCTCTTCAGGCTGTCAATGTTAAT |
| droEre2 |
scaffold_4845:19748460-19748547 + |
GATAAGCACGTTATTGTAAGCTGCGCC--ATA--TAGTTCGTAAA-------TTCGA--------------GCTTCAATTATGT---------T-----CTATTCTCTCCAGGCTGTCAATGTTAAT |
| droAna3 |
scaffold_12916:15881689-15881781 - |
GAAAAGCAAGTGATTGTAAGA--TGTAAAC---------------TCTTAGACCCTCCTAACTGCTACAA---GCTAATACTTT---------T-----GCTTTGAATCCAGGCTGTTAACGTGAAC |
| dp4 |
chr4_group3:8422191-8422279 + |
CAAAAGCAGCTGATAGTGAGTACTGGAAGC---------------ATTTATAT---------CTCTGGAAAGCACTAATTCTTT---------T-----TTTAATCTTCTAGGCTGTCGAATTAAAT |
| droPer1 |
super_1:9873274-9873362 + |
CAAAAGCAGCTGATAGTGAGTACTGGAAGC---------------ATTTATAT---------CTCTGGAAAGCACTAATTCTTT---------T-----TTGAATCTTCTAGGCTGTCGAATTAAAT |
| droWil1 |
scaffold_181132:892502-892596 - |
GACAAAGAAGTGATTGTAGGTGTCAATGA---------------T-------TTCA----------AACTAACATCAATTTTTTATTAACGTCTTTATTTTTTCCTATTCAGGCTATCAATGTCAAT |
| droVir3 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------- |