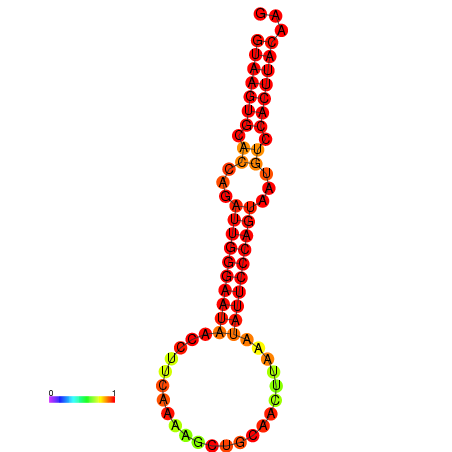
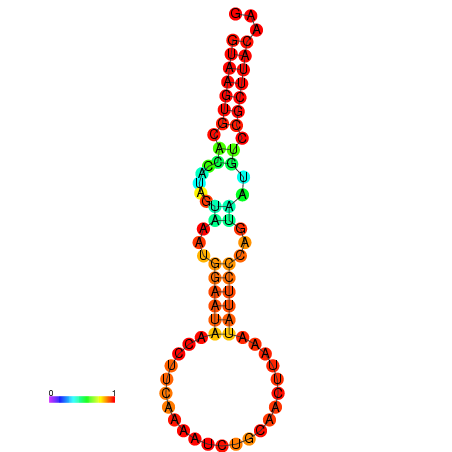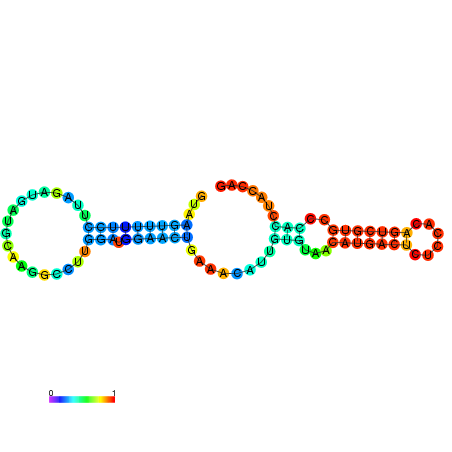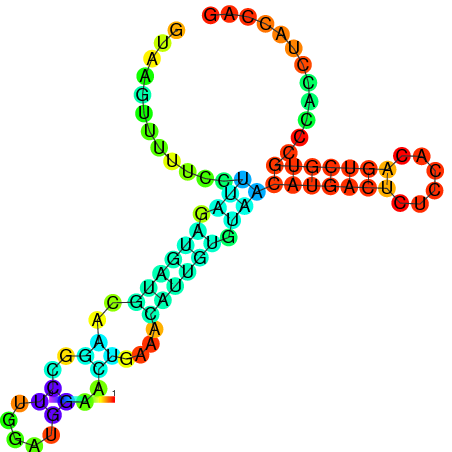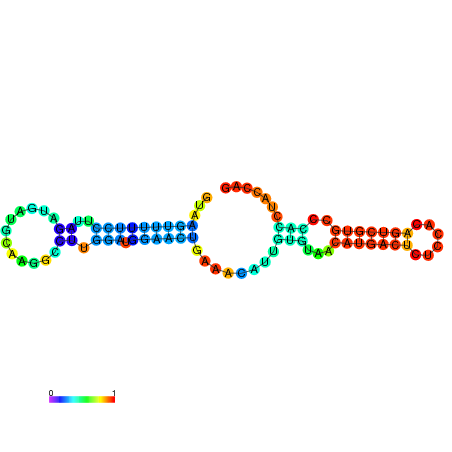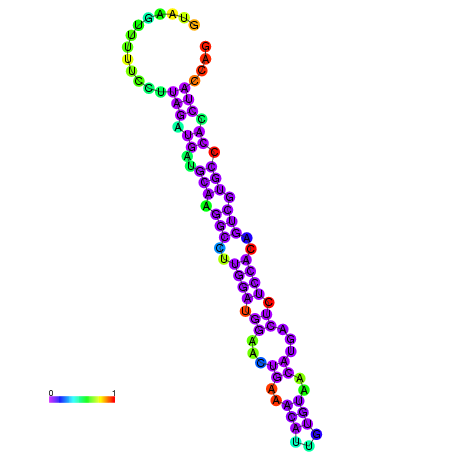| dm3 |
chr3R:12444473-12444619 + |
GGC--AACCTGCCACTGTCCATCTTCCGACCGGCGATCAGTAAGTGCACCAGATT------------G-GGAAT-----------------AACCTTCAAAAGCTGCAACTT-AAATATTCCCAGTAATGTCCACTTACAAGGAGCCCCTGGTTGGATGGGTGGACAA--CCTATTCGGACC |
| droSim1 |
chr3R:9022585-9022731 - |
GGC--AACCTGCCATTGTGCATCTTCCGACCGGCGATCAGTAAGTGCACCAGATT------------G-GGAAT-----------------AACCTTCAAAAGCTGCAACTT-AAATATTCCCAGTAATGTCCACTTACAAGGAGCCCCTGGTTGGATGGGTGGACAA--CCTATTCGGACC |
| droSec1 |
super_12:1702814-1702963 - |
GGC--ATCCTGCCATTGTGCATCTTCCGACCGGCGATCAGTAAGTGCACCATAGT-----A---A--ATGGAAT-----------------AACCTTCAAAATCTGCAACTT-AAATATTCCCAGTAATGTCCGCTTACAAGGAGCCCCTGGTTGGATGGGTGGACAA--CCTATTCGGACC |
| droYak2 |
chr3R:11948210-11948356 - |
GGC--GACCTGCCACTGTGCATCTTCCGCCCGGCGATCAGTAAGTGCACCAGATT------------G-GGATT-----------------AACCTTCAAAAGCTGCAGCTT-AACTATTCCCAGTAATGTCCACTTACAAGGAGCCCCTGGATGGCTGGGTGGACAA--CCTATTCGGACC |
| droEre2 |
scaffold_4770:9192977-9193123 - |
GGC--AACCTGCCCCTGTGCATCTTCCGCCCGGCGATCAGTAAGTGCAACAGATT------------G-GGAGT-----------------AACCGTCAAAAGCTACAACTT-CAATATTCCCAGTAATGTCCACTTACAAGGAGCCCCTGGTTGGCTGGATAGACAA--CCTATTCGGACC |
| droAna3 |
scaffold_13340:7435217-7435360 - |
GGC--AGCCTGCCAGTCAGCATTTATCGGCCTTCCATAAGTAAGTACTATATACG------------T-CCTGC-----------------TGTCTTATAAAATTGTTATTT----TTTTTCTAGTAATTGCCACATATAAGGAGCCATTAGTTGGATGGGTGGATAA--CCTTTATGGACC |
| dp4 |
chr2:5294042-5294203 - |
GGC--GACCTGCCCGTGTGCATTTTTCGGCCGGCCATTAGTAAGTTTTTCC----TTAGAT---G--AT-GCAAGGCCTTGGATGGAACTG-----AAACATTGTGTAACAT-GACTCTCCACAGTCGTGCCCACCTACCAGGAGCCCGTCGTTGGTTGGACTGACAA--TCTGTACGGGCC |
| droPer1 |
super_19:1006170-1006331 - |
GGC--GACCTGCCCGTGTGCATTTTTCGGCCGGCCATTAGTAAGTTTTTCC----TTAGAT---G--AT-GCAAGGCCTTGGATGGAACTG-----AAACATTGTGTAACAT-GACTCTCCACAGTCGTGCCCACCTACAAGGAGCCCGTCGTTGGTTGGACTGACAA--TCTGTACGGGCC |
| droWil1 |
scaffold_181108:4575766-4575914 - |
GGC--AATCTACCAATCTGTATTTTTCGCCCTGGTGTCAGTAAGTCCTCAAAATT------------C------------AACTTGTAACT-----ATAGCTGTTAAATTTTCTCCTTATTATAGTTATTGCCACCGCCAAGGAGCCAGTTTCGGGTTGGATTGACAA--TTTATATGGACC |
| droVir3 |
scaffold_13047:18500357-18500519 - |
ACAAAAAGCTGCCCATTATCGTCACAAGGCCTTCCATCGGTATGTAGGTCC----CGCT-A---A--CT-GGTTGGGCTTGACAAGTAACC-----ATATTTATTTTTTTTT-TTGTTCGCCCAGTGACGGCGGCCATACACGAGCCCATGCCCGGATGGATTGAGGG--CGTGAACGGACC |
| droMoj3 |
scaffold_6496:22335084-22335230 - |
GCC--GGTCTGCCGGCAGCCATAGTTAGGCCATCCATAGGTGAGTC----GAAGTATAAATCTAATTG-AG-AA-----------------GGCCCTAACTGACTGCTATT----------ACAGTATACGGTACACTGGAGCACCCCATGAAGGGCTGGGTGGGCAATGCCAACTCGGGTC |
| droGri2 |
scaffold_15074:333824-333971 - |
AGC--GGTTTGCCCATAAGCATCTTTCGCCCGGGTGTCAGTAAGTTCGTCGAATCATTAAGATCA----------------GTAAGAA---GATGTTCTCAGTCT-----------CCATTTCAGTTATTGGCAGCTACAAGGACCCCCTCCCGGGCTGGGTTGACAA--TCTATATGGTCC |