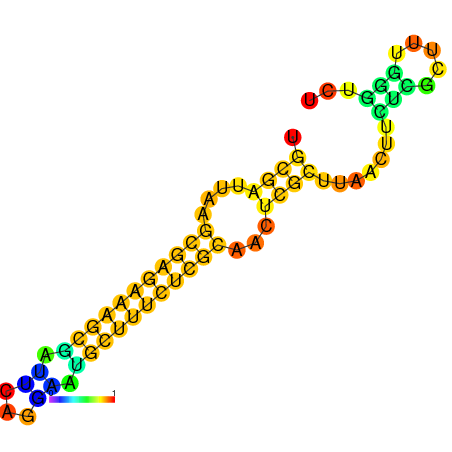
| dm3 |
chr2LHet:27321-27425 + |
ACAATCGTACGTTCTGAGCGAGTTGAAGCGATAAAGCAGGTTGCAAACTGCTCTCTCGCTTTGCTCTT---------------------------------------------------------ACTTTATCGCTTAAACTCGCA-----ACTCGCTTC----------GCTCTCA |
| droSim1 |
chr4:281112-281214 + |
AAAATCGTACGCTCTGAGCGAGTTGAAGCAATAAAGCAGGTTGCAAACTGCTCTTTCGCTTTGCTCTC---------------------------------------------------------GCTATATCGCTTGAGCTCGCA-----ACTCGCTTC----------GCT--CA |
| droSec1 |
super_30:85009-85111 + |
AAAATCGTACGCTCTGAGCGAGTTGAAGCAATAAAGCAGGTTGCAAACTGCTCTTTCGCTTTGCTCTC---------------------------------------------------------GCTATATCGCTTGAGCTCGCA-----ACTCGCTTC----------GCT--CA |
| droYak2 |
chrU:9061077-9061181 + |
ACAATCGTACGCTCTGAGCGAGTTGAAGCGATAAAGCAGGTTGCAAACTGCTCTCTCGCTTTGCTCTC---------------------------------------------------------GCTTTATCGCTTGAGCTCGCT-----ACTCGCCTC----------GCTCTCA |
| droEre2 |
scaffold_4784:23192819-23192904 - |
ACTATCAGACGCTCTGAGCGAGTTGCAGCGATAATGCAGGTTGCAAACTGTTCTCT----------------------------------------------------------------------------CGCTTGAGCTCGCA-----ACTCGTTTC----------GCTCTCA |
| droAna3 |
Unknown |
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| dp4 |
chr4_group4:3101286-3101457 + |
AGCAT---GAGC-GTCCACGAGATCAAGCGATAAGACATTTGCAAAGCTACTCTCTTGCTTCGCTTGCACTCGTTGCTGTCTTCGCTATAACTTGTTTCCCGCTCTTCACTGTAAATCGTTTTGCAC-TCTTCGCTCAAACTAGCCGCGCTGCTCTCTTCGCTTATCTTTGCTCTCA |
| droPer1 |
Unknown |
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 |
scaffold_181143:687820-687924 + |
ACACACGTATAACAGCAGCTGGATGAAGCGATAAAGCGA-TTCAGAATCGCTTTCTCGCAACTCGCTT------------------------------C--------------------------AACTTGTCGCTTTGGCTCTCT-----TCTCGCTTC----------GCTTGCA |
| droVir3 |
Unknown |
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6498:432956-433055 + |
AACAC---ACTTTAT-CATGCGATTAAGCGAGAAAGCGA-TTCAGGAATGCTTTCTCGCAACTCGCTT---------------------------------------------------------AACTTCTCGCTTTGGGTCTCT-----TCTCGCTTC----------GCTTGCA |
| droGri2 |
Unknown |
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |