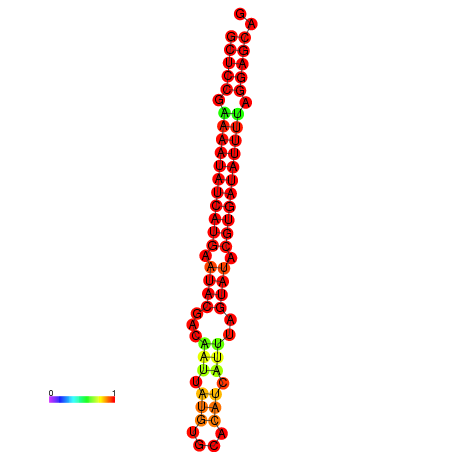
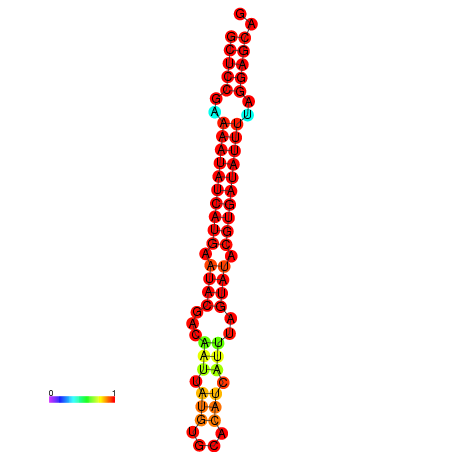
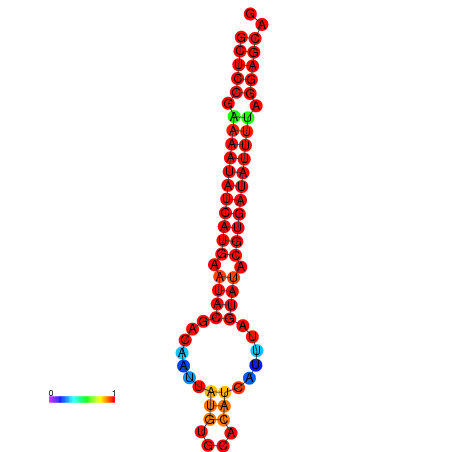
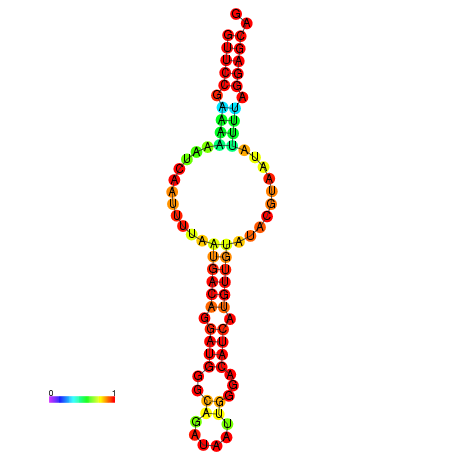
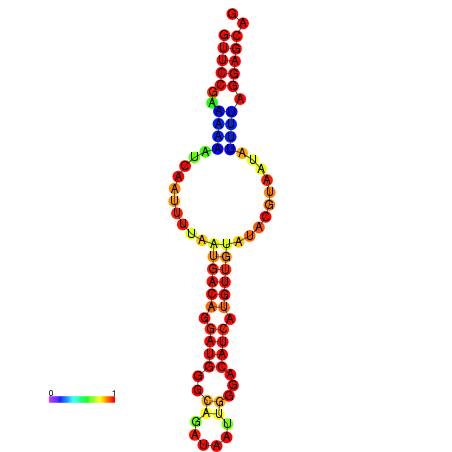
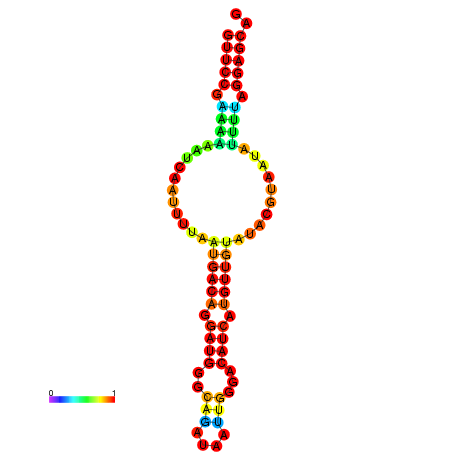
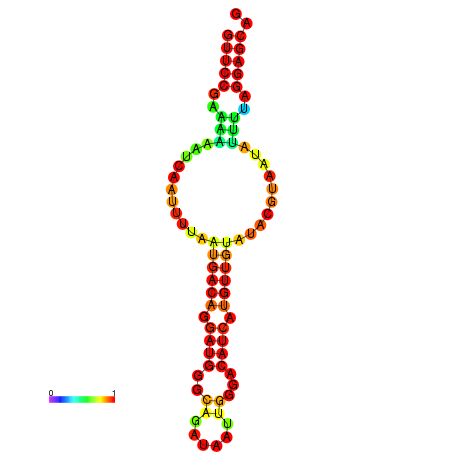
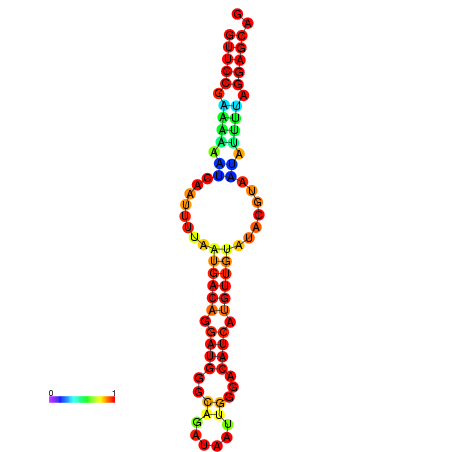
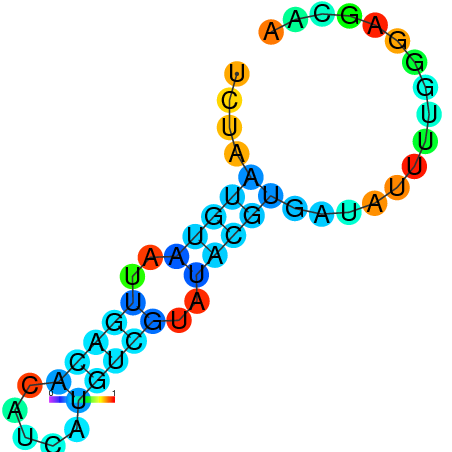
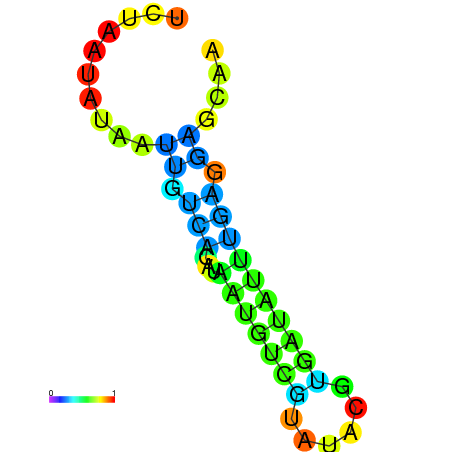
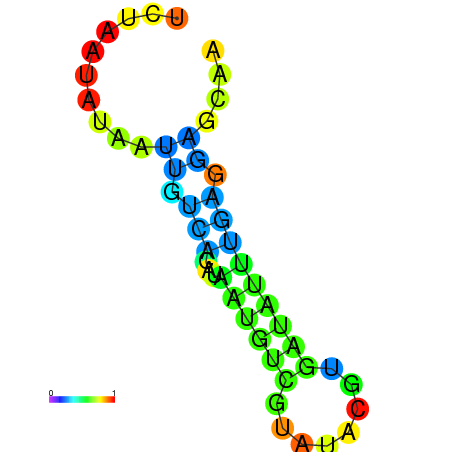
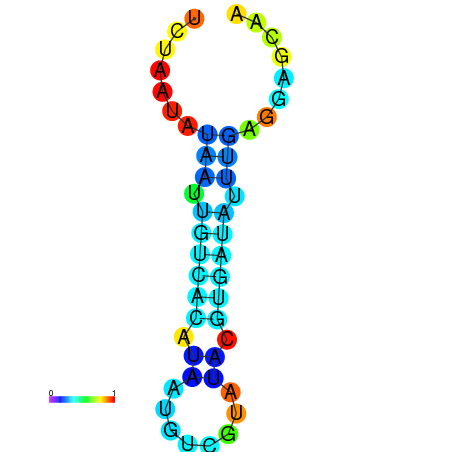| dm3 |
chr3R:5343231-5343336 + |
GAAAACTCACTCCTGAGCTGCTCCGAAAATATCATGAATA-CG----------ACAATTATGTGCACATCATTTAGTATACGTGATATTTTAGGAGCAGCTAAATTATATTTAAAGT |
| droSim1 |
wcy63a10.b1:18-120 - |
A---------------GTTGTTCCGAAAAAATCAATTTTAATGACAGGATGGGCAGATAATTGGGACATCATGTTGTATACGTAATATTTTAGGAGCAGCTCAATTATATTTAAAGT |
| droSec1 |
super_0:16553595-16553710 - |
GACAACTCACTCTTGAGCTGCTCCTAAAATATCACGTATA-CGACATGATGTCCCAATTACATGCACATCCTGTCGTATACGTGATATTTTAGGAGCAGCTCAATTATATTTAAAGT |
| droYak2 |
chr3R:9394463-9394539 + |
GGCAACCCAC--------------------------TCTAAT--------------GTAATTGACACATCATGTCGTATACGTGATATTTTGGGAGCAACGCAATTATATTTAAAGT |
| droEre2 |
scaffold_4770:16257712-16257788 - |
GGCAACTCAC--------------------------TCTAAT--------------ATAATTGTCACATAATGTCGTATACGTGATATTTGAGGAGCAAGTCAATTATATTTAAAGT |
| droAna3 |
Unknown |
--------------------------------------------------------------------------------------------------------------------- |
| dp4 |
Unknown |
--------------------------------------------------------------------------------------------------------------------- |
| droPer1 |
Unknown |
--------------------------------------------------------------------------------------------------------------------- |
| droWil1 |
Unknown |
--------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
Unknown |
--------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
Unknown |
--------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
Unknown |
--------------------------------------------------------------------------------------------------------------------- |