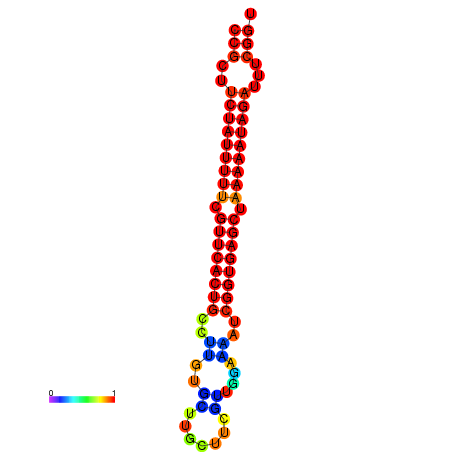
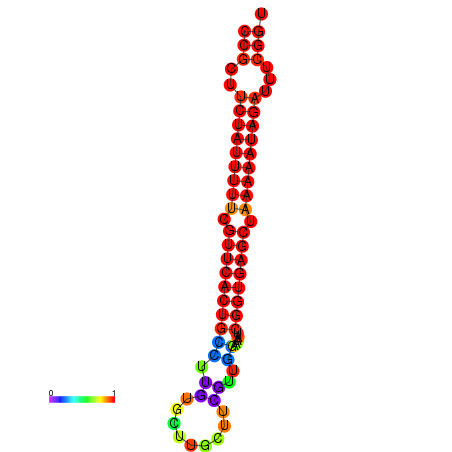
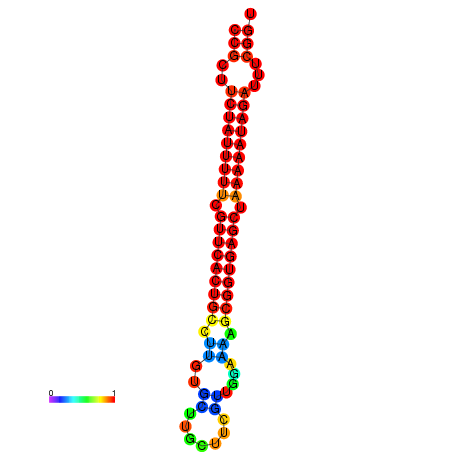
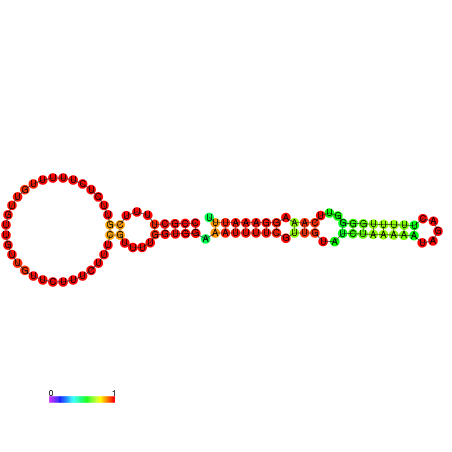
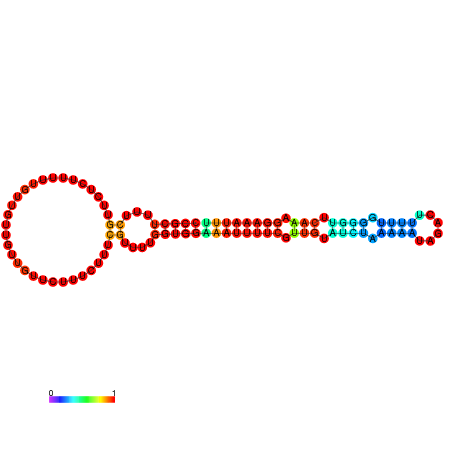
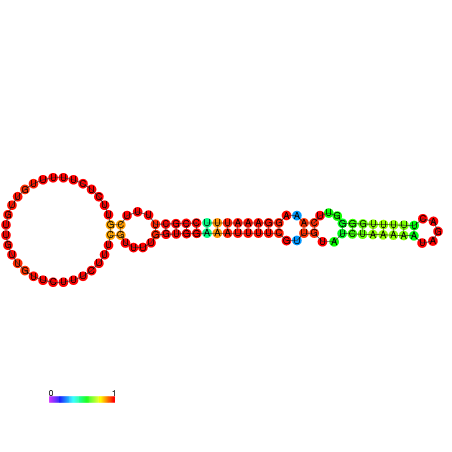
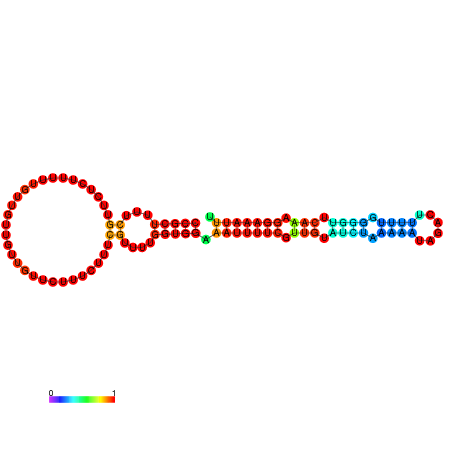
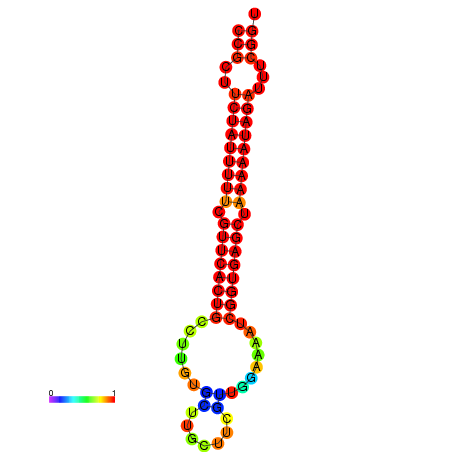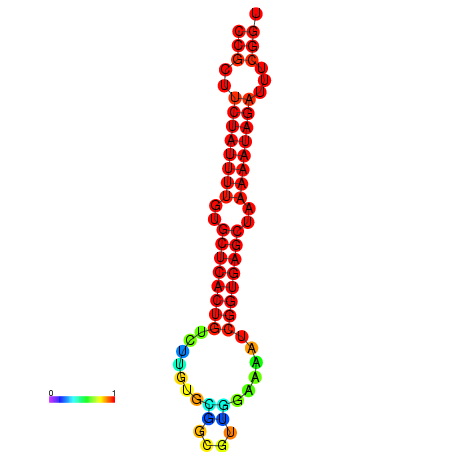| dm3 |
chr3L:3412130-3412266 - |
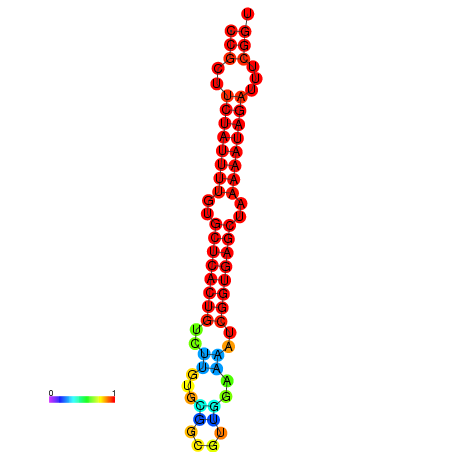
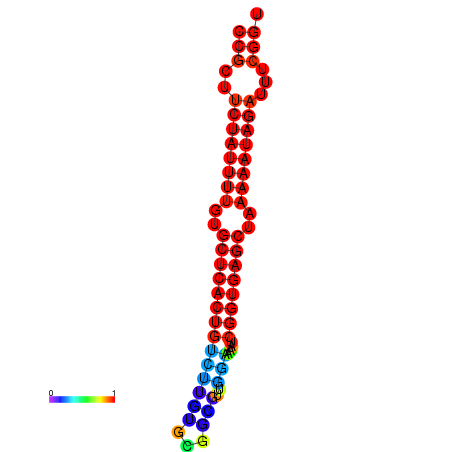
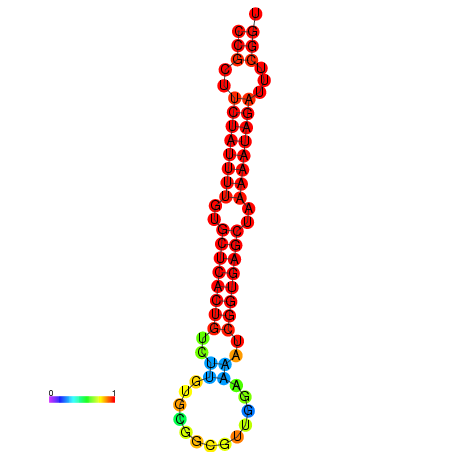
TGGGGCAGTGGAGCT-----G-----------------------------------------------------CCTGCAT---AACGC---------TGTGGCTTTCCGCTTCTATTTTTCGTTCACTGCCTTGTGC----TTCGTT--------------------------------------------------------------------------GGAAAATCGGTGAGCTAAAAATAGAATTCGGTTGCCACGCTTCAAGGGAAATGAGGAAAATTA |
| droSim1 |
zdo07f06.g1:327-468 + |
TGGGGCAGTGGAGCT-----G-----------------------------------------------------CCTGCAT---AAGGC---------TGTGGCTTTCCGCTTCTATTTTTCGTTCACTGCCTTGTGCTTGCTTCGTT--------------------------------------------------------------------------GGAAAATCGGTGAGCTAAAAATAGATTTCGGTTGCCACGCTTCAAGGGAAATGAGGAAAATTA |
| droSec1 |
super_2:3404661-3404801 - |
TGGGGCAGTGGAGCT-----G-----------------------------------------------------CCTGCAT---AAGGC---------TGTGGCTTTCCGCTTCTATTTTTCGTTCACTGCCTTGTGCTTGCTTCGTT--------------------------------------------------------------------------GGAAAAGCGGTGAGCTAAAAATAGATTTCGGTTGCCACGCTTCAAGGGAAATGAGGAAAATTA |
| droYak2 |
chr3L:3959855-3959989 - |
TGGGGCAGTGGGGCT-----G-----------------------------------------------------CAT--AT---AAGGC---------TGTGGCTTTCCGCTTCTATTTTGTGCTCACTGTCTTGTGC----GGCGTT--------------------------------------------------------------------------GGAAAATCGGTGAGCTAAAAATAGATTTCGGTTGCCACGCTTCGTGGAAAATGAGGAAAATTA |
| droEre2 |
scaffold_4784:6102142-6102274 - |
TGGGGCAGTGGAGCT-----G---------------------------------------------------------CAT---AAGGC---------TGTGGCTTTCCGCTTCTATTTTGTGTTCACTGTCCTGCGT----TTCGTT--------------------------------------------------------------------------GGAAAATCGGTGAGCCAAAAATAGATTTCGGTTGCCACGCTTCGAGGAAAATGCGGAAAATTA |
| droAna3 |
scaffold_13337:10031066-10031241 + |
TGGGGGCAGGTGGTG-----GCAGGAGGTGGCCGGAGGTGGTGGTAGGTGGGTGGGGGTGGTGTATGTAGGGGGCCTGTA------------------TGTGGCTTTCCGCTTCTACC--------ATTTTTTTTTGT-------------------------------------------------------TG----------------GAGGGTTCAGTGGAAATTCGGCGAGCTAAAAATATAATTCGGTTGCCACGCTTC---GGAAA----GAGAATTA |
| dp4 |
chrXR_group8:5510827-5511013 - |
TGTGTGTGTGTGTCT-----GT-----------------------GTGTGTGTGTGTGT-GTGTGTGTGT----GTGGCAGGGTGACAGCACTCTCTCTAAGGCTTTCCGCTTTTCGT------TCTCTTTT---TGT-TG-T-TGTTGTTCTTTCTTTCGTTTTGGTGGAAATTTTC----------GTTGTATCTAAAAATAGACTTTTTGGGGTTCAAAGGAAATTT-------------------------------------------TGTGGGACAAAA |
| droPer1 |
super_7263:1394-1409 + |
TACGGGTGTGAGACT-----G------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droWil1 |
scaffold_180955:1135974-1136099 - |
TATGGCGATATTTTGTAGGTT-----------------------------------------------------TTGGC-A---ATGGT---------TGTGGCTTTCCGCTTCTATTCAGT----------------------CGCA------------------CGAAAAATTTTCTCGGTTTTTTCCCAT-----------------------------------------GTCTAAAAATAGATTT------ACACCC---AAAGTA---GAAAAACATAA |
| droVir3 |
scaffold_13049:19660195-19660299 + |
G------------------------------------------------------------------------------------------------------TTCTGCGCTAGCTTGTTCGGTTTGGTAACATGAGC----TTCATT--------------------------------------------------------------------------AGTGTATGCAATAATGGCAAACATTTTTTGCCTGCCAGGCCTCGGCGAGGACGATGACGACGA |
| droMoj3 |
scaffold_6496:15371578-15371721 + |
TGCAACATTGATGAA-----A-----------------------------------------------------TATGCATAATGAAAATGTGCCCTCA-GAGTTATCCACG------------------------------------------------------------ATTTTTCTACTTTTTTTTTATACCCTGAAC---------CCATTTTCAATGGGAAATCAGGGTATA------ATGGTTTTGTTGAAATGTATG---TAACA----GGGAGAAG |
| droGri2 |
scaffold_15203:7704730-7704834 + |
CAGAATAATAGAATG-----T-----------------------------------------------------ACTCACT---TCAGC---------TATGGCATACAGATTTGCTTTATT-TGCTTTTCGTTGTAC-------------------------------------------------------------------------------------------------------------TTTCTGTTTTCTCAATGTACATGAAAAAGACTGAATTT |