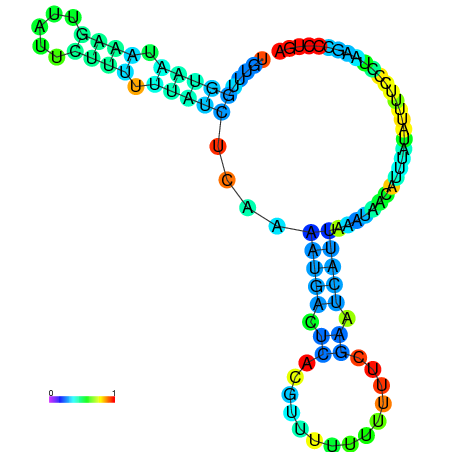
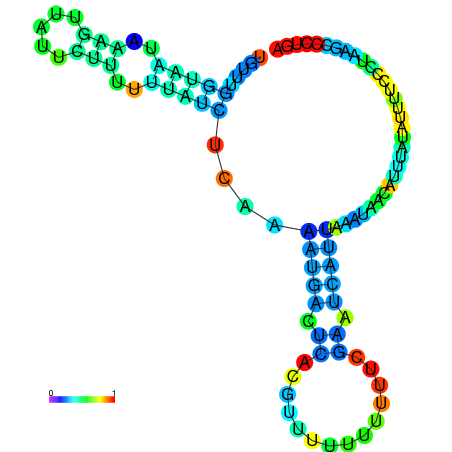
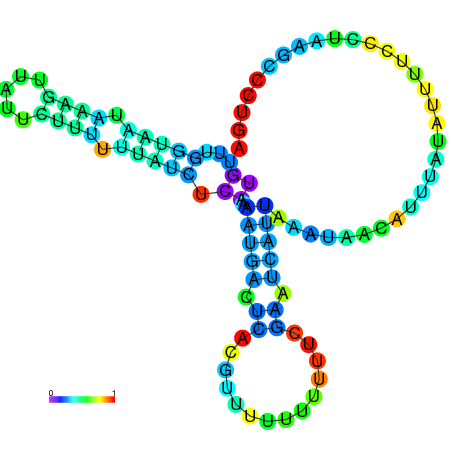
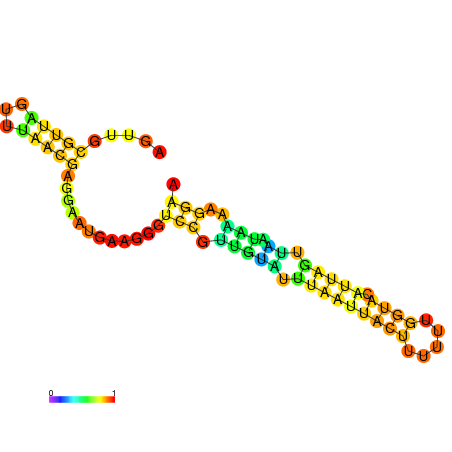
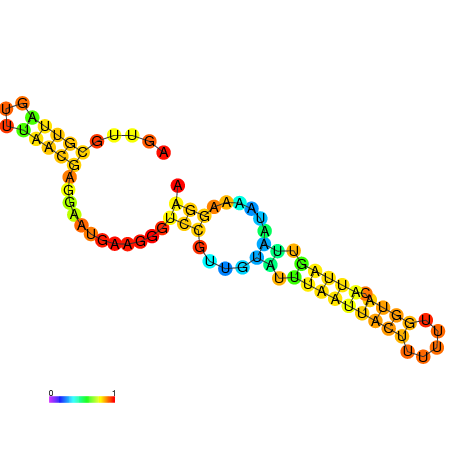
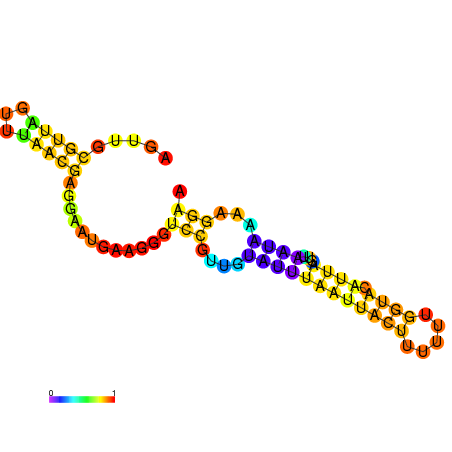
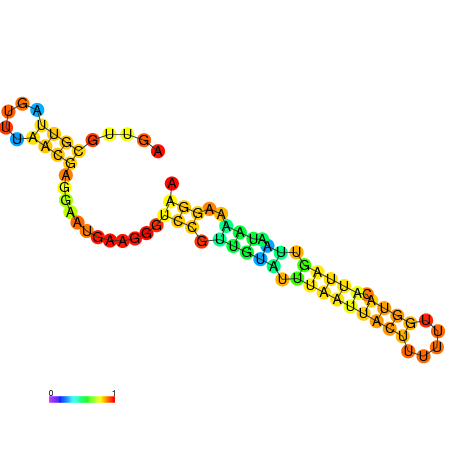
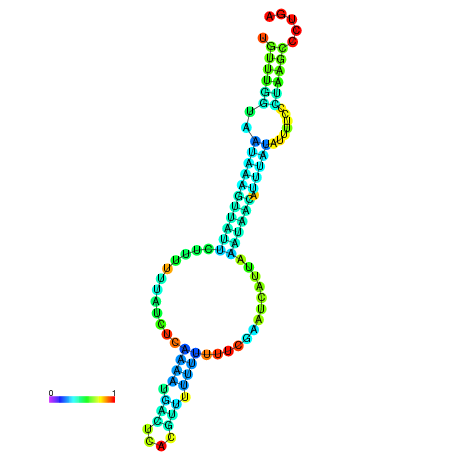| dm3 |
chr2L:1831670-1831814 - |
TGGAGAAGAGTGATATCGTTTAAGATGC---------TGTGAATATCCCGTTTTCTTAGCTTGGCAATAAAAT-ATTA------------------------------TTCACAGAAACTAGTAATTTACTT-----------CCAAGCTAACTGAAGGGGAT-ATTCAGTGCAACCACG--GACTAAAAATGCGTT-TG |
| droSim1 |
chr2L:1818710-1818856 - |
CGGAGCAGAGTGATATCGTTTGAGATGC---------TGTGAATATCCCGTTTTCTTAGCTTGGCAATAAAAT-ATTA------------------------------TTCACAGAAACTAGTAATTTACTT-----------CCAAGCTAACTGAAGGGGATTATTCAGTGCAACATCG--GACTAAAAATGCTTTTTG |
| droSec1 |
super_14:1778365-1778510 - |
CGGAGCAAAGTGATATCGTTTGAAATGC---------TGTGAATATCCCGTTTTCTTTGCTTGGCAATAAAAT-ATTA------------------------------TTCACAGAAACTAGTAACTTACTT-----------CCAAGCTAACTGAAGGGGAT-ATTCAGTGCAACCACG--GACTAAAAATGCTTTTTG |
| droYak2 |
chr2L:1804987-1805119 - |
CGGAGCAAACTGATGTCGTTTAGGATGC---------TGTGAATATCCCATTTTGTTAGCTTGGCAATAAAAT-ATTG------------------------------TTGATTGAAACTAGTAATTTACTT-----------CCAAGCTAACTGAAGTGGAT-ATTC--TA------------CAACAAATGGGTT-TT |
| droEre2 |
scaffold_4929:1878584-1878709 - |
CGGTG-----TGATTCTGCTTAAGGTGC---------TGTGAATATCCCATTTTCTTAGCTTGGCAATAAAAT-ATTA------------------------------TTCATCGA-ACTAGTAATTTAATT-----------CCAAGCCAACTAAAGGGGAT-ATTCACTG---------------CAAATGCATT-TG |
| droAna3 |
scaffold_12916:15757494-15757644 + |
CGTAGAAGCTT-AC-----TAGTTC--------------------------------TGTTTGGTAATAAAGT-TATTCTTTTTTATCTCAAAATGACTCACGTTTTTTTTTTCGA-ATCATTAAATAACATTTATATTTTCCCTAAGC-------CCTGAAT-ACTTATTATAAAAAAGACGATTATAAAAGTATA-TG |
| dp4 |
chr4_group3:8561836-8561913 - |
TGTTGTGGACTTAT-----CGGAGGAACTTGGCGAATTTTGATTATATTTTGATGCCATTTTT---------------------------------------------------------------------------------------ATCTGAAT-----------------------------AAAATGTTCT-TT |
| droPer1 |
super_1:10015828-10015916 - |
TGTTGTGGACTTAT-----CGGAGGAACTTGGCGAATTTTGATTATATTTAGATTCCATTTTT---------------------------------------------------------------------------------------ATCTGAATAAAAT-GTTC--TT---------------TAGTTACGTT-GA |
| droWil1 |
scaffold_180708:11841716-11841868 + |
TGCAGATGCCACAT-----CGGAATTGTGAAGGGAAAATGGGTTATTTAGTTGCGTTAGTTTAACGAGGAATGAAGGG------------------------------TCCGTTGTATTTAATTACTTTTTT----GGTAC--ATTAGTTAATAAAAGGAAAC-AATTAATCAATTAA----CATTATAAACTCGTT-TT |
| droVir3 |
Unknown |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
Unknown |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
Unknown |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |